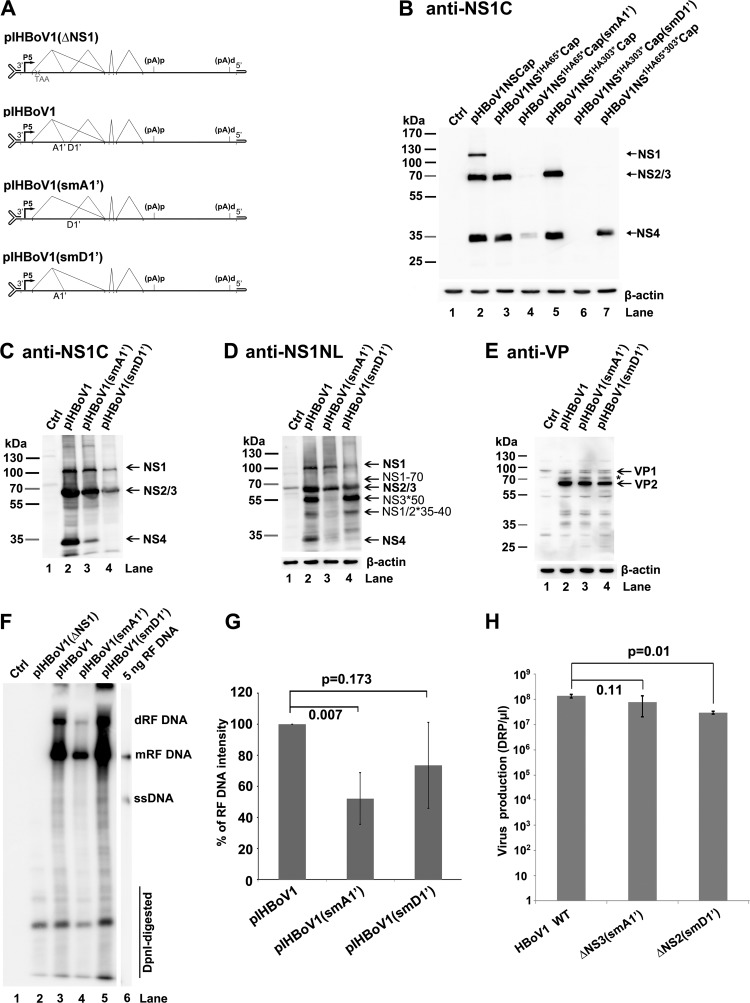
FIG 7.
Analyses of virus production from transfection of mutant HBoV1 infectious clones in HEK293 cells. (A) Diagrams of pIHBoV1 and its mutants. pIHBoV1(ΔNS1), which has a stop codon after NS1 aa 65 (11), and pIHBoV1(smA1′) and pIHBoV1(smD1′), in which the A1′ and D1′ splice sites, respectively, are knocked out, are diagramed, with transcription, splicing, and polyadenylation units shown. (B) NS knockdown efficiency from smA1′ and smD1′ mutations. HEK293 cells were transfected with plasmids with smA1′ and smD1′ mutations in the context of pHBoV1NSCap, as indicated. At 2 days posttransfection, cells were lysed for Western blotting using anti-NS1C antibody. (C to E) Western blot analysis of proteins from infectious clones. HEK293 cells were transfected with plasmids as indicated. Cells were harvested and lysed at 2 days posttransfection. The lysates were analyzed by Western blotting using anti-NS1C antibody (C) and reprobed with anti-NS1NL and anti-β-actin antibodies (D) and anti-VP antibody (E). The identities of detected proteins are shown with arrows at the right of the blot. The asterisk indicates a band that is likely a cleaved VP or an unknown VP. (F and G) Southern blot analysis of viral DNA replication. (F) HEK293 cells were transfected with plasmids as indicated. At 2 days posttransfection, cells were harvested for Hirt DNA preparations. The Hirt DNA samples were digested with DpnI and analyzed by Southern blotting with an HBoV1NSCap probe (19). The identities of detected bands are shown. dRF, double replicative form; mRF, monomer replicative form; ssDNA, single-stranded DNA. (G) Quantification of the RF bands, normalized to the level of DpnI-digested DNA, from three independent experiments is shown with means and standard deviations. (H) Quantification of virus production. HEK293 cells were transfected with plasmids as indicated. At 2 days posttransfection, cells were harvested for virus preparation. The final virus preparations (2 ml for each) were quantified for DNase I-resistant particles (DRP) using qPCR and are shown as DRP/μl on a log scale (y axis). Means and standard deviations are calculated from three independent experiments. P values are calculated using the Student t test.