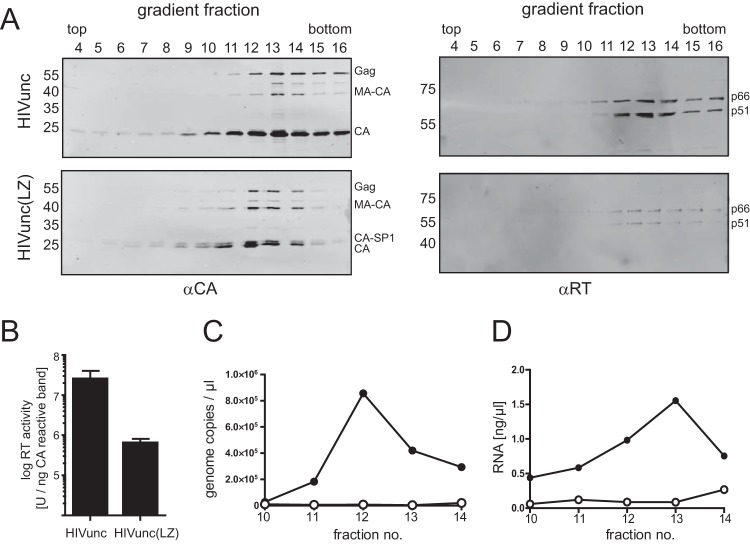
FIG 3.
Gradient purification of virus samples. VLPs were concentrated from the supernatant of HEK293T cells transfected with the indicated plasmids by ultracentrifugation through a 20% sucrose cushion and further purified by ultracentrifugation through an iodixanol density gradient. Fractions were collected from the top of the gradients and analyzed by immunoblotting (A), RT activity measurement (B), and RNA quantification (C, D). (A) Samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane. Viral proteins were detected by quantitative immunoblotting (LI-COR) using the indicated antisera. The positions of molecular mass standards (in kilodaltons) are marked at the left, and the positions of viral proteins are shown on the right. (B) The RT activity of gradient fractions was determined using a Sybr green product-enhanced RT assay (24). The values determined for peak fractions [fractions 13 and 12 for HIVunc and HIVunc(LZ), respectively] were normalized to the amount of anti-CA-reactive proteins detected in the respective fractions. The graph shows mean values and SDs from three independent experiments. (C, D) RNA content of peak fractions. RNA was extracted from gradient fractions 10 to 14 as described in Materials and Methods. Samples were analyzed for HIV-1-specific RNA by quantitative RT-PCR (C) or for total RNA content by staining with RiboGreen dye (D). Closed circles, HIVunc; open circles, HIVunc(LZ). Panels A, C, and D show data from one representative experiment out of three independent experiments.