Abstract
Premise of the study:
Sacred lotus (Nelumbo nucifera) is a perennial aquatic herbaceous plant of ecological, ornamental, and economic importance. MicroRNAs (miRNAs) play an important role in plant development. However, reports of miRNAs and their role in sacred lotus have been limited.
Methods:
Using the homology search of known miRNAs with genome and transcriptome contig sequences, we employed a pipeline to identify miRNAs in N. nucifera. We also predicted the targets of these miRNAs.
Results:
We found 106 conserved miRNAs in N. nucifera, and 456 of their miRNA targets were annotated. Quantitative real-time PCR (qRT-PCR) analysis revealed the different expression levels of the 10 selected conserved miRNAs in tissues of young leaves, stems, and flowers of N. nucifera. Negative correlation of expression level between five miRNAs and their target genes was also revealed.
Discussion:
Combining bioinformatics and experiment analysis, we identified the miRNAs in N. nucifera. The results can be used as a workbench for further investigation of the roles of miRNAs in N. nucifera.
Keywords: expressed sequence tag (EST), miRNA, Nelumbo nucifera, qRT-PCR, target genes
MicroRNAs (miRNAs) are ∼21-nucleotide noncoding small RNAs that play an important role in plant biological processes such as development, biotic and abiotic stress response, signal transduction, and protein degradation (Bartel, 2004). In plants, miRNA is transcribed by RNA polymerase II to primary miRNA transcripts called pri-miRNAs (Zhang et al., 2008). The pri-miRNAs are then cleaved to short stem-loop structured precursor miRNAs (pre-miRNAs) by the enzyme Dicer-like 1 (DCL1) and subsequently processed into an miRNA : miRNA* duplex (miRNA* is a small RNA on the opposite arm of the miRNA in the hairpin with partial complementarity to the miRNA). Single-stranded mature miRNA is then assembled into the RNA-induced silencing complex (RISC) and negatively regulates gene expression at the post-transcriptional level by targeting mRNA cleavage or inhibiting mRNA translation (Bartel, 2004; Zhang et al., 2008; Voinnet, 2009).
Currently, two strategies, including experimental and computational approaches, are widely used in miRNA identification. With regard to the experimental approaches, miRNA discovery can be achieved by direct cloning or deep sequencing from a small RNA library. Although both conserved and novel miRNAs can be identified with this approach, it was found to be tedious and expensive (Sunkar et al., 2005; Barrera-Figueroa et al., 2011). By contrast, the computer-based miRNA identification strategy employs a homology search, based on the principle that miRNAs are evolutionarily conserved (Zhang et al., 2006). After searching the public databases against the known miRNAs of the model plants or closely related species, miRNA homologs were obtained and predicted miRNAs were then verified on the basis of experimental validation (Zhang et al., 2008). The computational approaches are currently more popular in miRNA identification because they are fast, inexpensive, and effective. Thus, based on genomic survey sequence (GSS) and expressed sequence tag (EST) sequences in public data sets, conserved miRNAs in plants were identified, including Glycine max (L.) Merr. (Zhang et al., 2008), Nicotiana tabacum L. (Frazier et al., 2010), Solanum tuberosum L. (Xie et al., 2011), Brassica oleracea L. (Wang et al., 2012), cucurbit species (Hu et al., 2014), and other species.
Sacred lotus (Nelumbo nucifera Gaertn.) is a perennial aquatic herbaceous plant of ecological, ornamental, and economic importance (Hu et al., 2012). It is a source of herbal medicine because of strong antipyretic, cooling, astringent, antioxidant, anti-HIV, and demulcent properties. In Asia, N. nucifera has been widely cultivated for more than one thousand years (Shen-Miller et al., 2002). Due to its agricultural and medicinal significance, the whole genome of N. nucifera has been fully sequenced recently (Ming et al., 2013; Wang et al., 2013). However, reports of miRNAs and their roles in N. nucifera are limited. It is necessary to uncover miRNAs and target genes in N. nucifera. In this study, we aimed to identify miRNAs of N. nucifera and their target genes using homology-based computational approaches. To achieve this goal, we first performed a comparison between ESTs of N. nucifera and all known plant miRNA sequences. Then, a number of the putative miRNAs of N. nucifera were confirmed by expression analysis. Their relative expression level difference has been measured by quantitative real-time PCR (qRT-PCR) using miRNA-specific stem-loop RT and qRT-PCR primers (Varkonyi-Gasic et al., 2007).
MATERIALS AND METHODS
Sequence database and reference miRNAs
A total of 8316 known plant miRNAs were downloaded from the miRBase database (http://www.mirbase.org/; release 20, November 2013) (Kozomara and Griffiths-Jones, 2011) and used as a reference miRNA data set for identifying conserved miRNAs in N. nucifera. After removing the repeats, 2867 unique sequences were selected as the reference set. These miRNAs came from 24 plant species, including Aquilegia caerulea E. James, Arabidopsis lyrata (L.) O’Kane & Al-Shehbaz, A. thaliana (L.) Heynh., Brachypodium distachyon (L.) P. Beauv., Brassica napus L., B. rapa L., Camellia sinensis (L.) Kuntze, Glycine max, Gossypium hirsutum L., G. raimondii Ulbr., Hordeum vulgare L., Manihot esculenta Crantz, Medicago truncatula Gaertn., Nicotiana tabacum, Oryza sativa L., Populus trichocarpa Torr. & A. Gray, Prunus persica (L.) Batsch, Ricinus communis L., Solanum lycopersicum L., S. tuberosum, Sorghum bicolor (L.) Moench, Triticum aestivum L., Vitis vinifera L., and Zea mays L.
The 58,443 assembly genome contig sequences of the N. nucifera variety ‘China Antique lotus’ from both the National Center for Biotechnology Information (NCBI) database (accession: AQOG00000000.1) and RNA-seq data were used for miRNA mining (Ming et al., 2013). RNA-seq contigs_were generated for RNA-seq data from NCBI’s Short Read Archive (SRA) raw data (accession: SRX266474, SRX266489, SRX268456, and SRX265003) using the de novo assembly method from Trinity software (v2.0.2; Grabherr et al., 2011). The parameters used for assembly were as described (–CPU 12–kmer_method jellyfish 10G; Grabherr et al., 2011).
Identification of conserved miRNA and target prediction
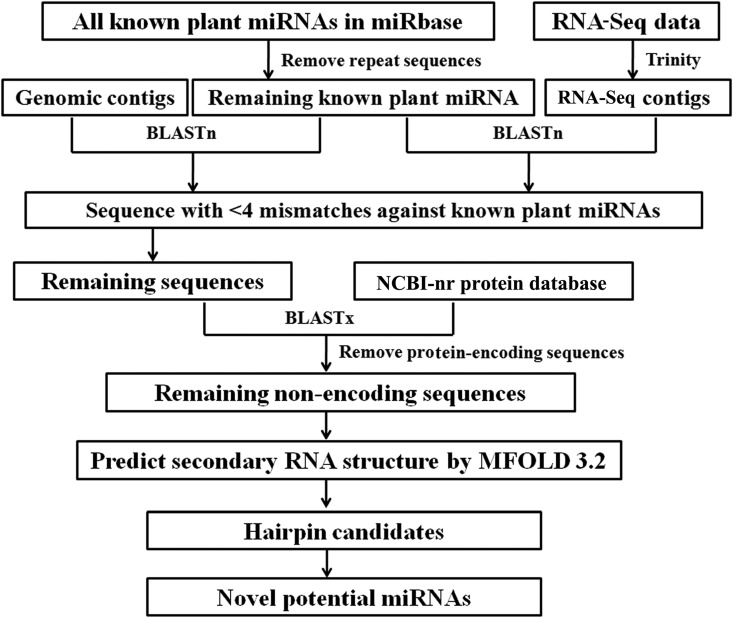
The genomic contig and RNA-seq contig sequences from the N. nucifera variety ‘China Antique lotus’ were used to mine conserved miRNA and their targets (Fig. 1). Computational identification of the conserved miRNA in sacred lotus was as described (Wang et al., 2012; Hu et al., 2014). Briefly, the N. nucifera genomic and RNA-seq contigs were aligned with known mature plant miRNAs using a BLASTn algorithm with an E threshold value of 10 and alignment length between 18 and 24 (Fig. 1). Results with no more than 3 nucleotide (<4 nucleotide) substitutions, including insertions, deletions, mutations, and gaps between known miRNAs and homolog sequences, were obtained from the BLASTn search (Wang et al., 2015). These alignment sequences to obtain the full-length sequences were extended from RNA-seq contig sequences. Protein-coding sequences were removed using BLASTx against NCBI’s nonredundant (nr) protein database. The secondary structures of the remaining sequences were predicted using MFOLD 3.2 software (Zuker, 2003). Finally, candidate miRNAs were identified based on the following criteria: (1) substitutions between contig sequences and known miRNA sequences not less than four; (2) minimum length of pre-miRNA = 45 nucleotides; (3) pre-miRNA can be folded into the perfect stem-loop hairpin secondary structure; (4) the miRNA : miRNA* duplex should have no loops; (5) mismatches in the miRNA : miRNA* duplex should not exceed 6 nucleotides; and (6) the negative minimal folding free energy (MFE) had the lower value and the minimal folding free energy index (MFEI) values of the predicted secondary structures should be higher than 0.85. In addition, psRNATarget was used to predict the targets of identified miRNAs with strict parameters, and the sequences were further compared against the NCBI-nr database for annotation (Dai and Zhao, 2011).
Fig. 1.
Workflow for identifying potential miRNAs in Nelumbo nucifera.
Validation of miRNA and the targets by qRT-PCR
For validation of miRNA and their targets in N. nucifera, young leaves, stems, and flowers of the N. nucifera accession of ‘Baihuajian lotus’ were selected and frozen immediately in liquid nitrogen and then stored at 80°C. Total RNA from each tissue sample was extracted using the RNAiso Reagent Kit (TaKaRa Bio Inc., Otsu, Shiga, Japan). Approximately 2 μg of each sample was reverse transcribed to stem-loop reverse transcription using specific RT primer and PrimeScript Reverse Transcriptase (TaKaRa Bio Inc.) for miRNAs (Varkonyi-Gasic et al., 2007; Quinn et al., 2015). Single-stranded cDNA for miRNA targets was synthesized using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. RT-PCR was then performed on an ABI StepOne Real-Time PCR System (Applied Biosystems, Carlsbad, California, USA) using SYBR Premix Ex Taq Kit (TaKaRa Bio Inc.). The melting curve was used to verify that only one specific product had been amplified. All the primers used are listed in Appendix S1 (92KB, xlsx) . The relative miRNA expression level was normalized and quantified using a ΔΔCT method. Three replicates were performed for each miRNA sample. Nelumbo nucifera NnEF1a (GI: 226897264) was used as the internal control for miRNA and their targets. Significant differences of the differential expression among different tissues in N. nucifera were evaluated by Student’s t test (*P < 0.05; **P < 0.01).
RESULTS
Identification of conserved miRNA and prediction of their targets
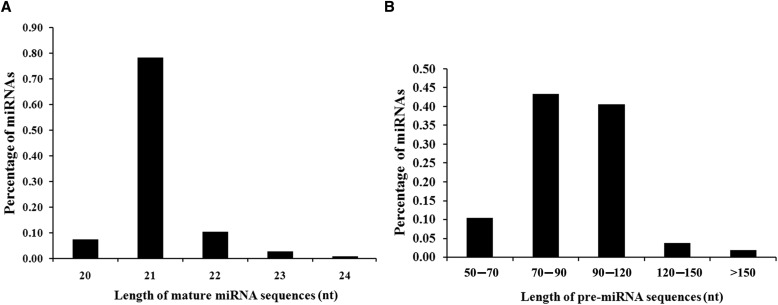
A total of 106 miRNAs, belonging to 40 families (Appendix S2 (92KB, xlsx) ), were identified with their secondary hairpin structure in this study. The length of mature miRNAs ranged from 20 to 24 nucleotides, with 21-nucleotide mature miRNAs being most abundant (83/106) (Fig. 2A). The length of pre-miRNAs ranged from 55 to 184 nucleotides, with an average length of 92 nucleotides (Fig. 2B).
Fig. 2.
Distribution of Nelumbo nucifera mature miRNA lengths (A) and pre-miRNA lengths (B).
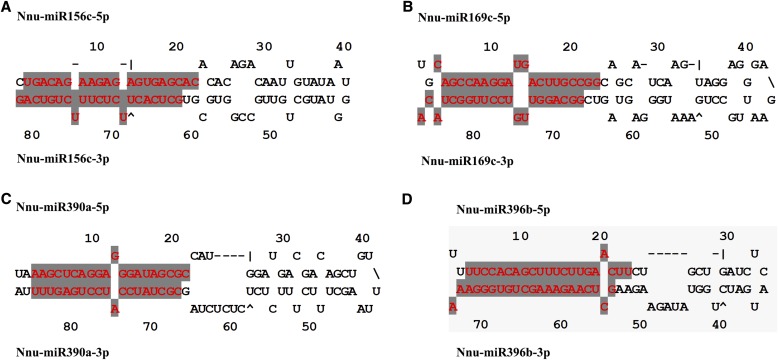
Several miRNA families were revealed in N. nucifera, including miR396 (13 loci), miR393 (7 loci), miR169 (6 loci), and miR172 (6 loci) (Appendix S2 (92KB, xlsx) ). Interestingly, 19 pairs of miRNA-5p/miRNA-3p were found to be located at the same precursors in this study (Fig. 3). Of them, the Nnu-miR396 family had the highest number of miRNA-5p/miRNA-3p pairs (Fig. 3D and Appendix S2 (92KB, xlsx) ). In particular, we found three pairs of sense and antisense miRNAs belonging to the same miRNA family (miR393) in N. nucifera (Fig. 4). The average value of MFE was −42.73 kcal/mol (range: −73.80 kcal/mol [Nnu-miR159a] to −13.60 kcal/mol [Nnu-miR5227]). The MFEI value of the predicted pre-miRNAs ranged from 0.62 (Nnu-miR2275b and Nnu-miR319a) to 1.51 (Nnu-miR396e-5p and Nnu-miR396e-3p), with an average of 1.01 (Appendix S2 (92KB, xlsx) ). Putative targets of these miRNAs were also predicted by psRNATarget. A total of 847 potential targets were identified for 40 miRNA families by searching for assembled RNA-seq sequences in N. nucifera. After aligning with the NCBI-nr protein database using BLASTx, 456 targets were annotated (Appendix S3 (92KB, xlsx) ).
Fig. 3.
Several miRNA-5p/miRNA-3p secondary structures in Nelumbo nucifera. (A) Nnu-miR156c-5p/3p; (B) Nnu-miR169c-5p/3p; (C) Nnu-miR390a-5p/3p; (D) Nnu-miR396b-5p/3p.
Fig. 4.
Sense and antisense miRNAs and their secondary structures in Nelumbo nucifera.
Expression analysis of miRNA and their targets using qRT-PCR
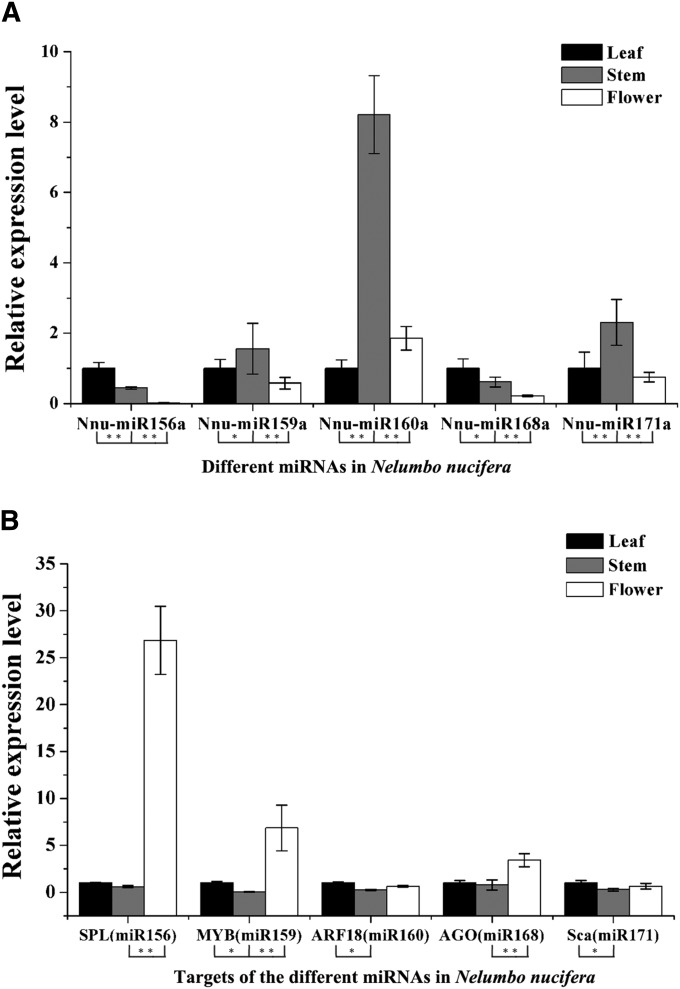
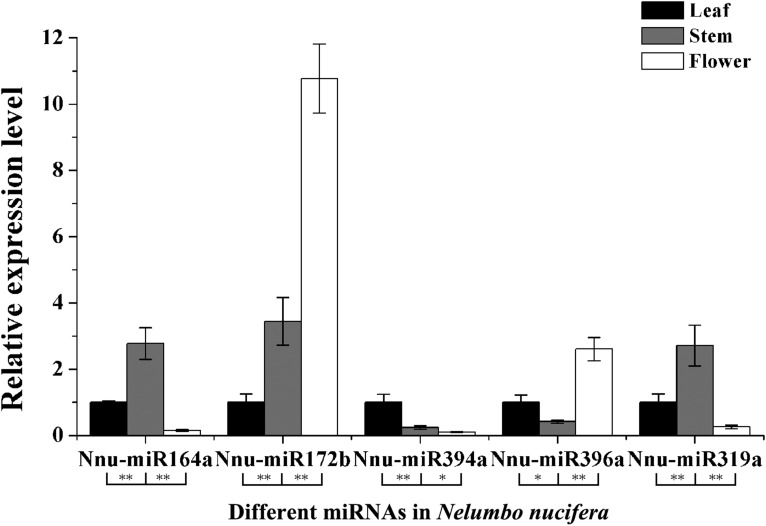
To confirm the expression of miRNAs in N. nucifera, we selected 10 miRNAs (miRNAs 156, 159, 160, 164, 168, 171, 172, 319, 394, and 396) using qRT-PCR (Table 1 and Appendix S1 (92KB, xlsx) ). Of them, five miRNAs (miRNAs 156, 159, 160, 168, 171) and their targets were further analyzed to validate the expression level. The 10 miRNAs exhibited differential expression patterns, thus revealing expression differences among N. nucifera leaf, stem, and flower tissues (Figs. 5, 6).
Table 1.
The 10 selected conserved miRNAs for qRT-PCR analysis in Nelumbo nucifera.
| miRNA family | GenBank accession no. | Target protein |
| Nnu-miR156a | comp21843_c0_seq1 | Squamosa promoter-binding-like protein 16 |
| Nnu-miR159a | comp12092_c0_seq1 | Transcription factor GAMYB-like |
| Nnu-miR160a | comp17106_c0_seq1 | Auxin response factor 18-like |
| Nnu-miR164 | comp11773_c0_seq1 | NAC domain-containing protein 100-like |
| Nnu-miR168a | comp17877_c0_seq1 | Argonaute1-1 (AGO1-1) |
| Nnu-miR171a | comp16457_c0_seq1 | Scarecrow-like protein 27-like |
| Nnu-miR172 | comp9596_c0_seq1 | APETALA2 (AP2) |
| Nnu-miR319 | comp12092_c0_seq1 | Transcription factor GAMYB-like |
| Nnu-miR394 | comp11701_c0_seq1 | F-box only protein 6 |
| Nnu-miR396 | comp3877_c0_seq1 | Growth-regulating factor 3 isoform 2 |
The miRNA and its target gene were analyzed by qRT-PCR.
Fig. 5.
The relative expression levels of five miRNAs and target genes in young leaves, stems, and flowers of Nelumbo nucifera. (A) The relative expression levels of five miRNAs in N. nucifera. (B) The relative expression levels of five corresponding target genes in N. nucifera. Significant differences of the differential expression among different tissues in N. nucifera were evaluated by Student’s t test (*P < 0.05; **P < 0.01).
Fig. 6.
The relative expression levels of five miRNAs in young leaves, stems, and flowers of Nelumbo nucifera. Significant differences of the differential expression among different tissues in N. nucifera were evaluated by Student’s t test (*P < 0.05; **P < 0.01).
Moreover, the target gene expression patterns of the five selected miRNA demonstrated a negative association with their miRNAs in N. nucifera stem and flower tissue (Fig. 5). For example, the expression level of miR156 was found to be lowest in N. nucifera flower tissue, while the highest expression level of the miR156 target gene (SPL) was observed in the same tissue. Similar results were also found in the four other miRNAs and their target genes (Fig. 5).
DISCUSSION
Currently, few studies have been reported to identify miRNAs and their targets with experimental validation in N. nucifera. In this study, we identified a total of 106 miRNAs in N. nucifera, belonging to 40 families. Furthermore, three pairs of antisense miR393 were found in N. nucifera. Antisense miRNA is a class of miRNA that is transcribed from both sense and antisense transcripts derived from the same genomic loci; some of the antisense miRNAs were complementary to their sense miRNAs (Fig. 4). This antisense miRNA has been found in plants, such as Brassica oleracea, Cucumis melo L., and Cucumis sativus L. (Wang et al., 2012; Hu et al., 2014). The antisense miRNA potentially targets the genes that differ from their sense partners, although the evolution and function of the antisense miRNA is still unknown.
In this study, all of the 106 identified miRNAs have been predicted to target 456 genes. After annotation, these genes were likely to play essential roles in growth and development as well as in other biological processes. Furthermore, some conserved miRNAs and their targets were validated in N. nucifera by qRT-PCR (Figs. 5, 6). These include:
• Nnu-miR156 targets SPL, which were negatively regulated in post-transcriptional expression. It has been well documented that miR156 in plants plays a critical role for vegetative to reproductive-phase transition (Poethig, 2009). In this study, we also indicated that Nnu-miR156-targeted SPLs are involved in flower development in N. nucifera.
• Nnu-miR159 targets GAMYB-like transcription factor. The interaction between miR159 families and phytohormones has been demonstrated in various plant responses. In Arabidopsis, miR159 expression is regulated by gibberellin (GA) and abscisic acid (ABA) during anther development (Achard et al., 2004). In strawberry, miR159 families interact with GAMYB in relation to changes in GA content during receptacle development. In ornamental gloxinia (Sinningia speciosa (Lodd.) Hiern), the expression level of miR159 was negatively correlated with GAMYB, which was effectively involved in flowering time control during flower development (Li et al., 2013). Similarly, we observed that Nnu-miR159 may participate in flower development of N. nucifera.
• Nnu-miR160 targets the auxin response factor 18-like (ARF18-like) in N. nucifera. In Arabidopsis, miR160 negatively modulates three ARF genes (ARF10/ARF16/ARF17), controlling root cap development and primary and lateral root growth (Wang et al., 2005). Similarly, miR160/ARFs governs root and nodule organogenesis in Medicago truncatula (Bustos-Sanmamed et al., 2013). We have found that Nnu-miR160 potentially targets the ARFs to regulate stem development in N. nucifera.
• Nnu-miR168 targets ARGONAUTE1 (AGO1) that encodes the RNA slicer enzyme of miRNAs. AGO1 interacts with miR168 to regulate the small RNA pathway in Arabidopsis. It is also reported that miR168 plays crucial roles in many development processes in tomato, including phase transition, leaf epinasty, and fruit development (Xian et al., 2014). In this study, we showed that Nnu-miR168 regulates flower development in N. nucifera.
• Nnu-miR171 targets SCARECROW-LIKE 27 (SCL27). In previous studies, miR171-targeted scarecrow-like proteins (SCL6/SCL22/SCL27) have been revealed to negatively regulate chlorophyll biosynthesis (Wang et al., 2010; Curaba et al., 2013). In Arabidopsis, miR171 interacts with scarecrow-like proteins to combine GT cis-elements, modulating gibberellin-regulated chlorophyll biosynthesis under light conditions (Ma et al., 2014). However, in N. nucifera, Nnu-miR171-targeted genes may play a role during stem development.
• On the other hand, five of the 10 miRNAs (Nnu-miR164, Nnu-miR172, Nnu-miR394, Nnu-miR396, and Nnu-miR396) have been measured with relative expression levels by qRT-PCR.
• miR164 targets NAC, NAM (no apical meristem), ATAF (Arabidopsis transcription activation factor), and CUC (cup-shaped cotyledon) transcription factors. The NAC genes contain a complementary site with miR164, which were negatively regulated by miR164 through mRNA cleavage (Sieber et al., 2007). In Arabidopsis, miR164 is also involved in age-dependent cell death leaves (Kim et al., 2009). In recent reports, the functions of miR164 have been proven to participate in the responses of plants to biotic as well as abiotic stress (Fang et al., 2014).
• miR172 targets APETALA2 transcription factor (Jung et al., 2011). miR172 is critical for the control of flowering time and floral morphology. It is reported that over-expression of miRNA172 leads to early flowering and suppresses the floral organ specification in Arabidopsis (Aukerman and Sakai, 2003).
• miR394 targets gene LCR (LEAF CURLING RESPONSIVENESS) that encodes an F-box protein (SKP1-Cullin/CDC53-F-box). Besides its control of stem cell competence in the Arabidopsis shoot meristem (Knauer et al., 2013), miR394-regulated LCR abundance plays a role in fine-tuning their responses to ABA and abiotic stress (Song et al., 2013).
• miR396 targets GROWTH-REGULATING FACTORs (GRFs), a plant-specific family of transcription factors (Hewezi et al., 2012). Previous studies have demonstrated that miR396-targeted AtGRFs are required for mediating cell differentiation during leaf morphogenesis in Arabidopsis (Mecchia et al., 2013).
• miR319 regulates transcription factors of the TCP family, controlling leaf morphogenesis and several other plant developmental processes in Arabidopsis and tomato (Schommer et al., 2012).
Conclusions
In this study, using in silico computer-based approaches for identifying miRNAs and their target genes based on a sequence homology search, 106 conserved miRNAs, belonging to 40 families, were identified in N. nucifera. Pre-miRNAs varied from 55 to 184 nucleotides in length, while mature miRNAs ranged from 20 to 24 nucleotides. Four hundred fifty-six potential miRNA targets were also predicted in this study. Negative correlation of the expression levels between five miRNAs (miRNAs 156, 159, 160, 168, and 171) and their target genes were observed in leaves, stems, and flowers of N. nucifera. Our study results can be used as a workbench for further study of N. nucifera miRNAs and those in other plant species.
Supplementary Material
LITERATURE CITED
- Achard P., Herr A., Baulcombe D. C., Harberd N. P. 2004. Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365. [DOI] [PubMed] [Google Scholar]
- Aukerman M. J., Sakai H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Figueroa B. E., Gao L., Diop N. N., Wu Z. G., Ehlers J. D., Roberts P. A., Close T. J., et al. 2011. Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biology 11: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bustos-Sanmamed P., Mao G., Deng Y., Elouet M., Khan G. A., Bazin J., Turner M., et al. 2013. Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Functional Plant Biology 40: 1208–1220. [DOI] [PubMed] [Google Scholar]
- Curaba J., Talbot M., Li Z., Helliwell C. 2013. Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biology 13: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhao P. X. 2011. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Research 39(Suppl 2): W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Xie K., Xiong L. 2014. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. Journal of Experimental Botany 65: 2119–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier T. P., Xie F. L., Freistaedter A., Burklew C. E., Zhang B. H. 2010. Identification and characterization of microRNAs and their target genes in tobacco (Nicotiana tabacum). Planta 232: 1289–1308. [DOI] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T., Maier T. R., Nettleton D., Baum T. J. 2012. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiology 159: 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. H., Pan L., Liu H. G., Wang S. Z., Wu Z. H., Ke W. D., Ding Y. 2012. Comparative analysis of genetic diversity in sacred lotus (Nelumbo nucifera Gaertn.) using AFLP and SSR markers. Molecular Biology Reports 39: 3637–3647. [DOI] [PubMed] [Google Scholar]
- Hu J. H., Sun L. L., Zhu Z. X., Zheng Y., Xiong W., Ding Y. 2014. Characterization of conserved microRNAs from five different cucurbit species using computational and experimental analysis. Biochimie 102: 137–144. [DOI] [PubMed] [Google Scholar]
- Jung J. H., Seo P. J., Kang S. K., Park C. M. 2011. miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Molecular Biology 76: 35–45. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Woo H. R., Kim J., Lim P. O., Lee I. C., Choi S. H., Hwang D., Nam H. G. 2009. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Knauer S., Holt A. L., Rubio-Somoza I., Tucker E. J., Hinze A., Pisch M., Javelle M., et al. 2013. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Developmental Cell 24: 125–132. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. 2011. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research 39(Suppl 1): D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Bian H., Song D., Ma S., Han N., Wang J., Zhu M. 2013. Flowering time control in ornamental gloxinia (Sinningia speciosa) by manipulation of miR159 expression. Annals of Botany 111: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Hu X., Cai W., Huang W., Zhou X., Luo Q., Yang H., et al. 2014. Arabidopsis miR171-targeted scarecrow-like proteins bind to GT cis-elements and mediate gibberellin-regulated chlorophyll biosynthesis under light conditions. PLOS Genetics 10: e1004519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecchia M. A., Debernardi J. M., Rodriguez R. E., Schommer C., Palatnik J. F. 2013. MicroRNA miR396 and RDR6 synergistically regulate leaf development. Mechanisms of Development 130: 2–13. [DOI] [PubMed] [Google Scholar]
- Ming R., VanBuren R., Liu Y. L., Yang M., Han Y. P., Li L. T., Zhang Q., et al. 2013. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biology 14: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R. S. 2009. Small RNAs and developmental timing in plants. Current Opinion in Genetics & Development 19: 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn C. R., Rie I., Danilo D. F. 2015. Computational predictions and expression patterns of conserved microRNAs in loblolly pine (Pinus taeda). Tree Genetics & Genomes 11: 806. [Google Scholar]
- Schommer C., Bresso E. G., Spinelli S. V., Palatnik J. F. 2012. Role of microRNA miR319 in plant development. In R. Sunkar (ed.), MicroRNAs in plant development and stress responses, 1st ed. Springer, Berlin, Germany. [Google Scholar]
- Shen-Miller J., Schopf J. W., Harbottle G., Cao R. J., Ouyang S., Zhou K. S., Southon J. R., Liu G. H. 2002. Long-living lotus: Germination and soil γ-irradiation of centuries-old fruits, and cultivation, growth, and phenotypic abnormalities of offspring. American Journal of Botany 89: 236–247. [DOI] [PubMed] [Google Scholar]
- Sieber P., Wellmer F., Gheyselinck J., Riechmann J. L., Meyerowitz E. M. 2007. Redundancy and specialization among plant microRNAs: Role of the MIR164 family in developmental robustness. Development 134: 1051–1060. [DOI] [PubMed] [Google Scholar]
- Song J. B., Gao S., Sun D., Li H., Shu X. X., Yang Z. M. 2013. miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid–dependent manner. BMC Plant Biology 13: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Girke T., Jain P. K., Zhu J. K. 2005. Cloning and characterization of microRNAs from rice. Plant Cell 17: 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R., Wood M., Walton E. F., Hellens R. P. 2007. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687. [DOI] [PubMed] [Google Scholar]
- Wang H. Y., Xiao S. H., Wang M., Kim N. H., Li H. X., Wang G. L. 2015. In silico identification of conserved microRNAs and their targets in bovine fat tissue. Gene 559: 119–128. [DOI] [PubMed] [Google Scholar]
- Wang J. W., Wang L. J., Mao Y. B., Cai W. J., Xue H. W., Chen X. Y. 2005. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Y., Yang X. D., Xu H. B., Chi X. Y., Zhang M., Hou X. L. 2012. Identification and characterization of microRNAs and their target genes in Brassica oleracea. Gene 505: 300–308. [DOI] [PubMed] [Google Scholar]
- Wang L., Mai Y. X., Zhang Y. C., Luo Q., Yang H. Q. 2010. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Molecular Plant 3: 794–806. [DOI] [PubMed] [Google Scholar]
- Wang Y., Fan G. Y., Liu Y. M., Sun F. M., Shi C. C., Liu X., Peng J., et al. 2013. The sacred lotus genome provides insights into the evolution of flowering plants. Plant Journal 76: 557–567. [DOI] [PubMed] [Google Scholar]
- Xian Z., Huang W., Yang Y., Tang N., Zhang C., Ren M., Li Z. 2014. miR168 influences phase transition, leaf epinasty, and fruit development via SlAGO1s in tomato. Journal of Experimental Botany 65: 6655–6666. [DOI] [PubMed] [Google Scholar]
- Xie F. L., Frazier T. P., Zhang B. H. 2011. Identification, characterization and expression analysis of microRNAs and their targets in the potato (Solanum tuberosum). Gene 473: 8–22. [DOI] [PubMed] [Google Scholar]
- Zhang B. H., Pan X. P., Wang Q. L., Cobb G. P., Anderson T. A. 2006. Computational identification of microRNAs and their targets. Computational Biology and Chemistry 30: 395–407. [DOI] [PubMed] [Google Scholar]
- Zhang B. H., Pan X. P., Stellwag E. J. 2008. Identification of soybean microRNAs and their targets. Planta 229: 161–182. [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.