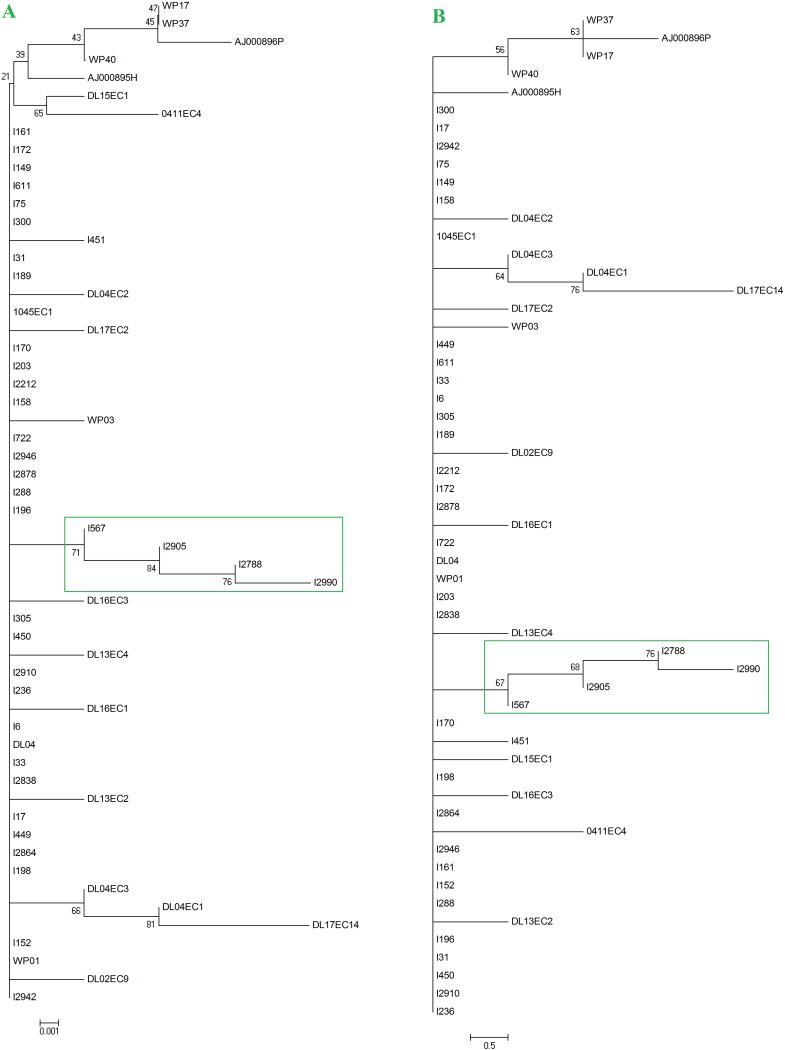
Fig. 1.
Phylogenetic comparison of our study isolates with the global Ascaris population: Internal transcribed spacer 1 (ITS1) sequences of our study isolates were aligned with ITS1 sequences from the global Ascaris population using ClustalW multiple alignment program of MEGA version 4 software. Two separate phylogenetic trees were generated from this alignment using two individual methods- [A] Neighbor-Joining (NJ) and [B] Maximum parsimony (MP). In both cases, widely distributed and most prevalent Ascaris genotype G1 (Genbank ID AJ554036) was considered as out-group. The bootstrap values were also analyzed to estimate confidence intervals. Phylogenetic comparison using these two individual methods (i.e. NJ and MP) generates trees with similar topology. In both cases, few of our Indian isolates (with newly identified ITS1 sequences) formed distinct cluster with high bootstrap value (marked with green color in both trees), which may indicates their distinct phylogenetic position in respect to global Ascaris population.