Figure 3.
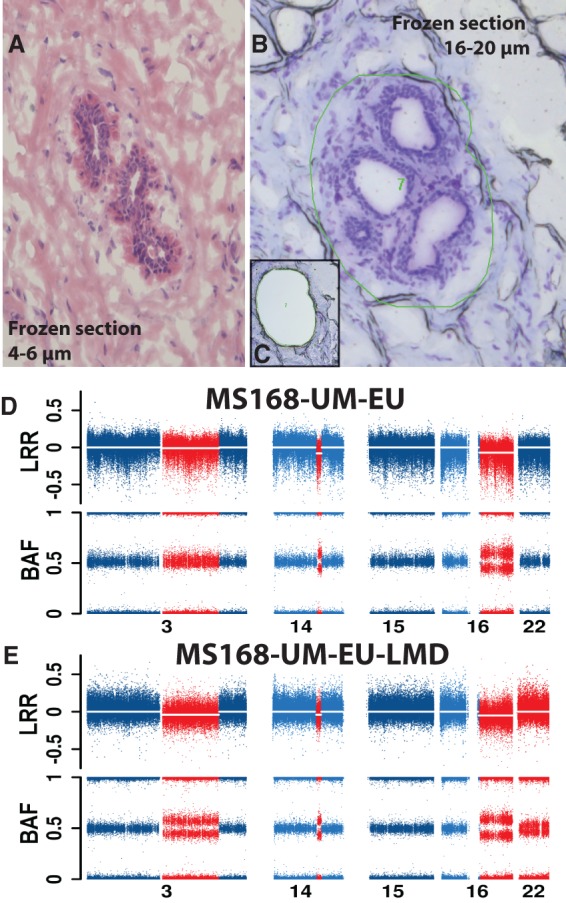
Laser-microdissection (LMD) validation of three deletions on Chromosome 3, 14, and 16 in normal cells from sample MS168-UM-EU. (A) A representative image of normal breast parenchyma (hematoxylin and eosin staining) in thin frozen section from specimen MS168-UM-EU, with a normal duct. (B,C) Images before and after the normal structures have been dissected by laser and collected. The thick frozen sections (16–20 µm) in B and C have been stained with cresyl violet. The green irregular circle in B shows the area marked for dissection by laser. (D,E) Genetic copy number profiles of chromosomes with aberrations (in red) and without (in blue) from SNP arrays. The profile in D has been produced using the bulk DNA derived from all cells in sample MS168-UM-EU, while the profile in E is derived from DNA isolated from microdissected cells. Sample MS168-UM-EU shows deletions present in ∼5%–15% of cells, as indicated by the BAF values deviating from the value of 0.5. The corresponding number of cells affected by deletions in sample MS168-UM-EU-LMD is higher, suggesting an enrichment of cells with aberrations. The combined load of deletions on Chromosomes 3, 14, and 16 in the sample MS168-UM-EU is 92.8 Mb. Interestingly, the microdissected sample MS168-UM-EU-LMD contains also a low proportion of cells (∼5%–10%) with a copy number neutral loss of heterozygozity (CNNLOH) of whole Chromosome 22, which was not detectable in the bulk DNA derived from all cells in sample MS168-UM-EU.
