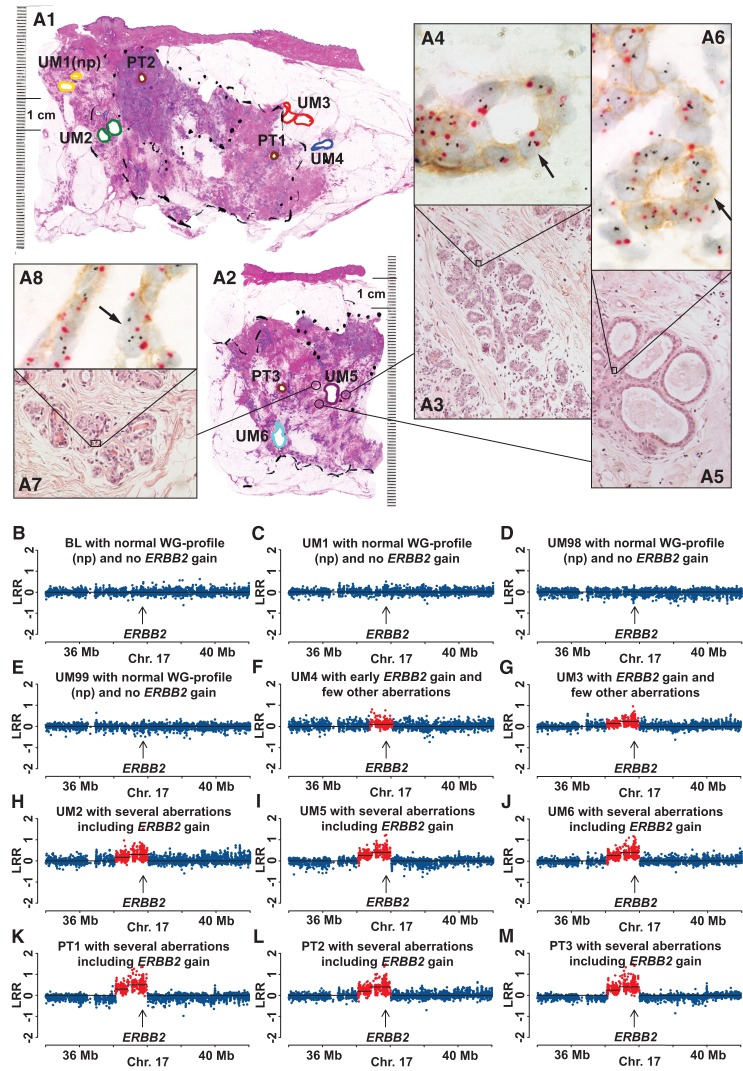
Figure 6.
Multimodal examination of pathology, gene copy number, and gene expression for case AW020, with evidence of increased copy number and expression of ERBB2 in normal epithelial cells. (A1,A2) Two section levels of large-format histology slides of breast tissue stained with hematoxylin and eosin, with diagnosis of multifocal invasive ductal carcinoma (Luminal B, HER2+). Areas of tissue samples taken for DNA extraction, prior to formalin fixation of the tissue, are marked with colored thick lines. Positions of three primary tumors 1, 2, and 3 (PT1, PT2, and PT3) are shown in brown. In A1, UM1, UM2, UM3, and UM4 are labeled in yellow, green, red, and blue, respectively. In A2, UM5 and UM6 are labeled in purple and light blue, respectively. (np) Normal genetic profiles (see also below, B–M). Three cores from paraffin-embedded tissue (thin-lined black circles) surrounding sample UM5 were taken for separate analysis using the HER2 tricolor Dual ISH DNA Probe Cocktail Assay (Roche) and the results are shown in A3–A8. (A3,A5,A7) Histological images of normal breast tissue with cross-sections through terminal ductal lobular units (TDLUs). Black arrows in A4, A6, and A8 point to single nuclei of normal epithelial cells containing more than two copies of ERBB2 (black dots). The centromere of Chromosome 17 is stained in red. Note a weak but clearly discernible immunohistochemical staining of HER2 protein in the cell membrane of normal epithelial cells upon high magnification. (B–M) A segment of Chromosome 17 containing ERBB2 in 12 samples from Illumina global genome analysis. Blood (BL, normal control tissue), UM1, UM98, and UM99 samples have normal profiles (np) with no gain of ERBB2. The remaining eight samples were scored as containing an increased copy number (red dots) for ERBB2. The samples UM98 and UM99 are taken from parts of breast tissue as far away as possible from the segment (lobe) affected by breast cancer and are not visualized in A1 and A2. The total size of aberrations in UM samples is as follows: UM4, 0.7 Mb; UM3, 7.4 Mb; UM2, 191 Mb; UM5, >39% of the genome; UM6, >39% of the genome.