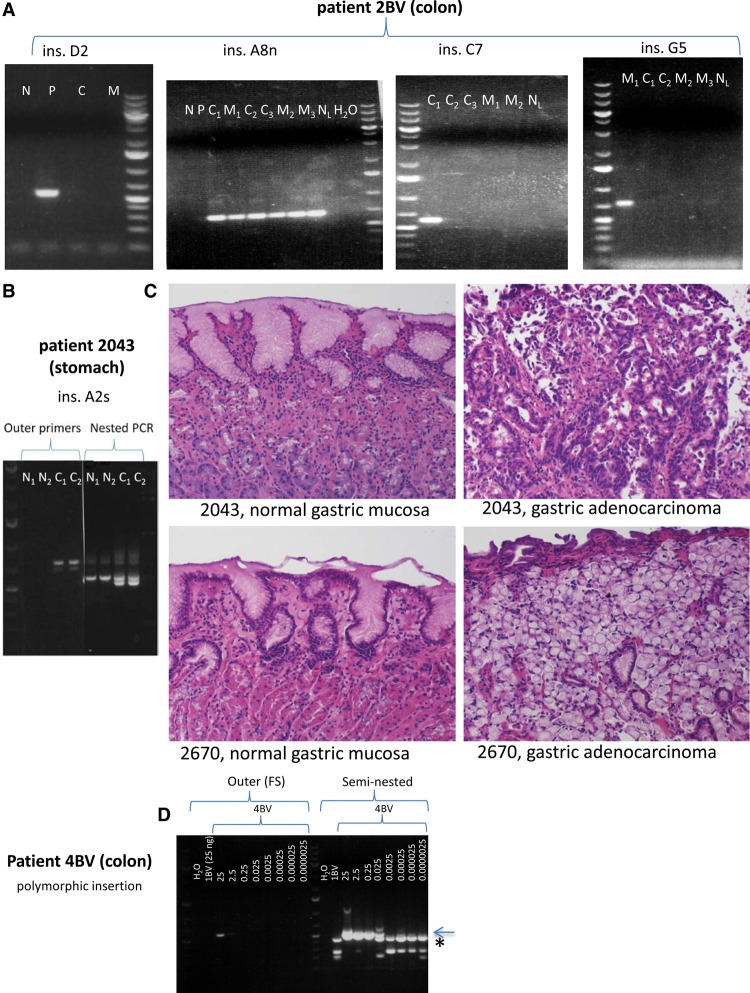
Figure 3.
Representative examples of further somatic L1 insertions in colon cancer patient 2BV and gastric cancer patient 2043. (A) L1 insertions in colorectal cancer (all detectable by conventional PCR), from left to right: ins. D2 (polyp-specific, present exclusively in adenoma sample “5”); ins. A8n (cancer-and-metastasis-specific, present in three independent primary colorectal cancer sections and in three independent liver metastasis sections); ins. C7 (the only insertion in colon cancer cases that is detectable in only one tumor section, “6/1”); ins. G5 (a metastasis-specific insertion, present in only one of three sections of metastasis sample “7”). (N) Normal colon, (P) polyp, (C) primary colorectal cancer, (C1) primary colorectal cancer section 1, (C2) primary colorectal cancer section 2, (C3) primary colorectal cancer section 3, (M) metastasis, (M1) metastasis section 1, (M2) metastasis section 2, (M3) metastasis section 3, (NL) normal liver. (B) The fifth agarose gel shows the second somatic normal-and-cancer-specific L1 insertion in stomach cancer patient 2043 (detectable in cancer both by conventional and nested PCR, but detectable in normal stomach exclusively by nested PCR). (N1) Normal stomach section 1, (N2) normal stomach section 2, (C1) primary gastric cancer section 1, (C2) primary gastric cancer section 2. (C) Hematoxylin and eosin (H&E) staining revealed normal gastric mucosa in case of insertions A2s and C1s (Fig. 2C). (Top, left) Normal gastric mucosa from patient 2043; (top, right) gastric adenocarcinoma with intestinal differentiation from patient 2043. (Bottom, left) Normal gastric mucosa from patient 2670; (bottom, right) gastric adenocarcinoma with signet ring features from patient 2670. All photomicrographs were taken at 20× original magnification. H&E-stained slides were sectioned and stained in the Johns Hopkins Department of Pathology according to standard protocols. (D) In order to estimate the limits of detection of an insertion by conventional and seminested PCR, we performed a dilution series on DNA containing a heterozygous polymorphic germline insertion. Normal colon DNA containing a polymorphic L1 insertion of patient 4BV was mixed with normal colon DNA of patient 1BV, which did not contain this insertion, and served as competitor DNA. The amount of DNA used for conventional PCR in the case of 4BV DNA is shown in ng, which is constituted to a final amount of 25 ng using 1BV DNA. One microliter of each FS PCR product was used for seminested PCR. The detection limit of an insertion using conventional PCR appears to be 2.5 ng DNA against 22.5 ng competitor DNA (faint PCR band present). However, using seminested PCR, the detection limit is 25 pg DNA against 24.975 ng competitor DNA. Sanger sequencing confirmed that the lower molecular weight seminested PCR bands are nonspecific (seen in 1BV and in 4BV using ≤0.025 ng DNA for conventional PCR, and is marked by an asterisk), while the higher molecular weight band is the correct PCR product (marked by arrow). Thus, we detect one copy in 10 cells (very faint band) with conventional PCR, and one copy per 1000 cells with seminested PCR. Identical results were obtained for a second heterozygous polymorphic L1 and a different competitor DNA. O'GeneRuler 1 kb Plus DNA ladder was used (Thermo Scientific).