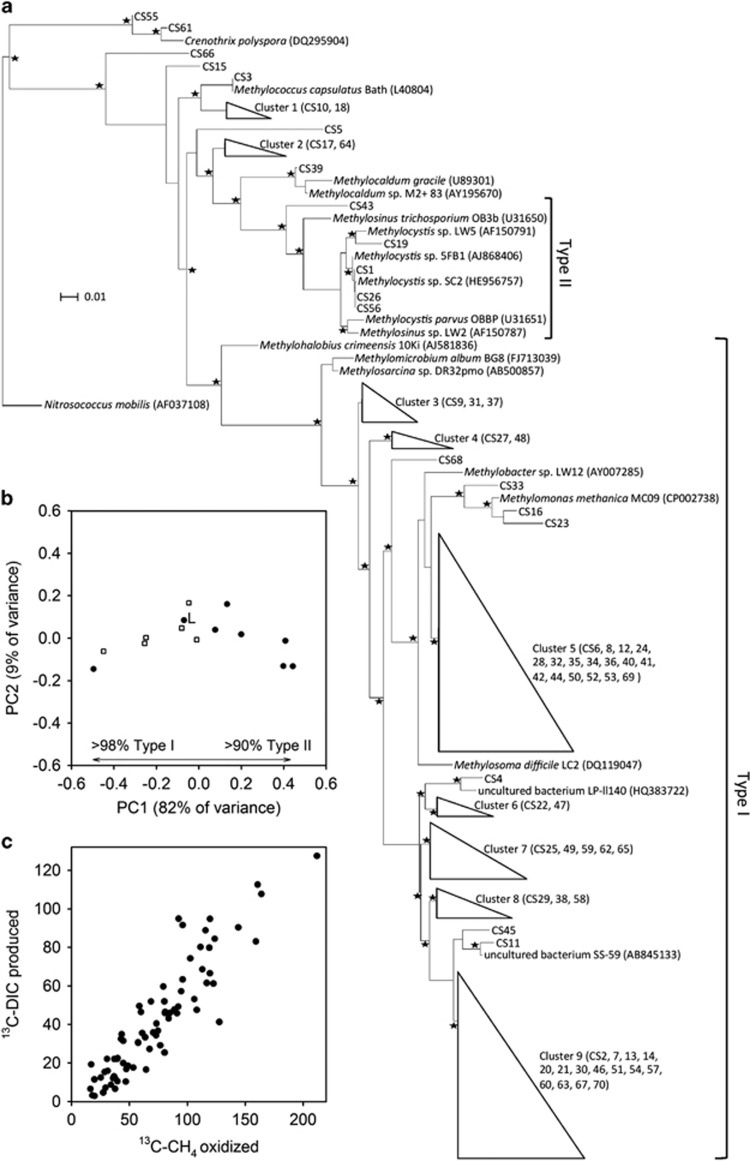
Figure 5.
A measure of the variation in the methanotroph community and overall CCE for a sample of eight chalk streams in southern England. (a) Maximum likelihood tree of representative pmoA sequences (about 200 bp). Collapsed branches are indicated by triangles. The amoA gene of Nitrosococcus mobilis (AF037108) was used as the out group. Asterisks indicate local support values over 75%. The bar represents 0.01 average nucleotide substitutions per base. (b) Principal coordinates analysis of the methanotroph community based on pmoA sequences. Filled circles are the eight independent stream samples and open squares the intrastream variation in the Lambourn. L is the pooled sample from the Lambourn also used to measure its CCE. (c) Standardized output from the mixed-effect model showing the best estimate of the overall ratio of DIC produced per CH4 consumed (slope 0.50, s.e. 0.03, t 16.4) and CCE via methanotrophy (1−0.50 × 100) was 50% on average for the eight rivers (filled circles in b).