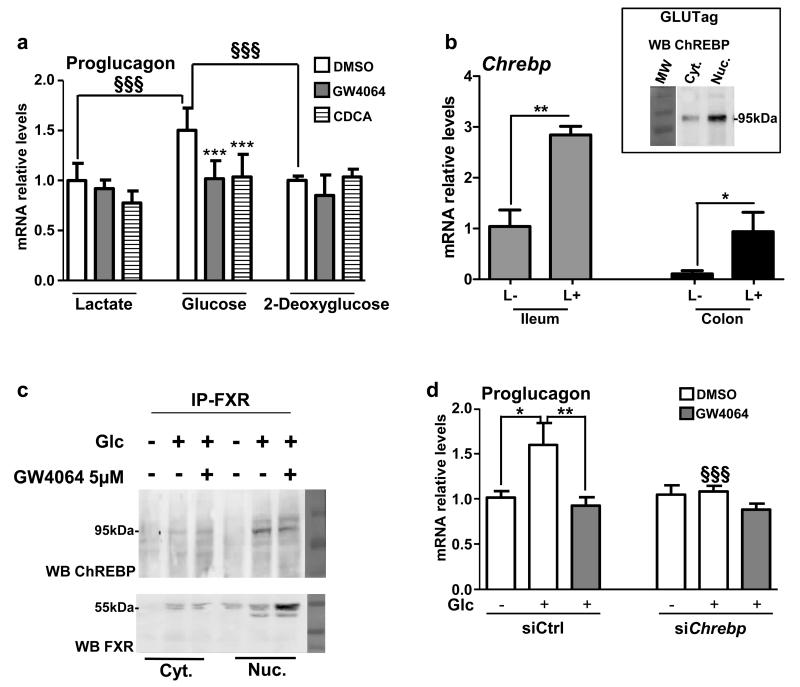
Figure 4. FXR inhibits glucose-induced proglucagon expression.
(a) Proglucagon qPCR on cDNA from GLUTag cells starved for 12h with lactate (10 mmol L−1) and then incubated for 24h in lactate (10 mmol L−1), glucose (5.6 mmol L−1) or 2-deoxyglucose (5.6 mmol L−1) media containing DMSO, GW4064 (5 μmol L−1) or CDCA (100 μmol L−1) (n=3; performed 3 times). Data are represented as mean +/− SD. Two-Way ANOVA analysis followed by Bonferronni’s posthoc test. ***P≤0.001: effect of GW4064 and CDCA on proglucagon mRNA levels in each medium conditions. §§§P≤0.001: effect of glucose on proglucagon mRNA levels in DMSO, GW4064 and CDCA conditions. (b) Chrebp qPCR on cDNA from FACS-sorted proglucagon-negative and proglucagon-positive cells from the ileum (ileum L−; ileum L+) and colon (colon L−; colon L+) of GLU-VENUS mice (lower panel; n=3) and ChREBP protein expression from cytoplasm and nucleus extract from GLUTag cells (upper panel; performed 3 times). Data are represented as mean +/− SD. Student t-test. *P≤0.05 and **P≤0.01 (c) ChREBP and FXR western-blots after FXR immunoprecipitation on lysates from cytoplasm and nucleus of GLUTag cells treated or not with GW4064 (5 μmol L-1) in presence or not of glucose (5.6 mmol L−1) (performed 2 times). (d) Proglucagon qPCR on cDNA from GLUTag cells electroporated with a siCtrl or siChrebp, starved for 12h with lactate (10 mmol L−1) and then incubated for 24h in lactate 10 mmol L−1 (Glc −) or glucose 5.6 mmol L−1 (Glc +) media supplemented with DMSO or GW4064 (5 μmol L−1) (n=3; performed 3 times). Data are represented as mean +/− SD. Two-Way ANOVA analysis followed by Bonferronni’s posthoc test. *P≤0.05 and **P≤0.01: effect of treatments on each transfection condition. §§§P≤0.001: effect of siChrebp in each treatment condition.