Figure 1.
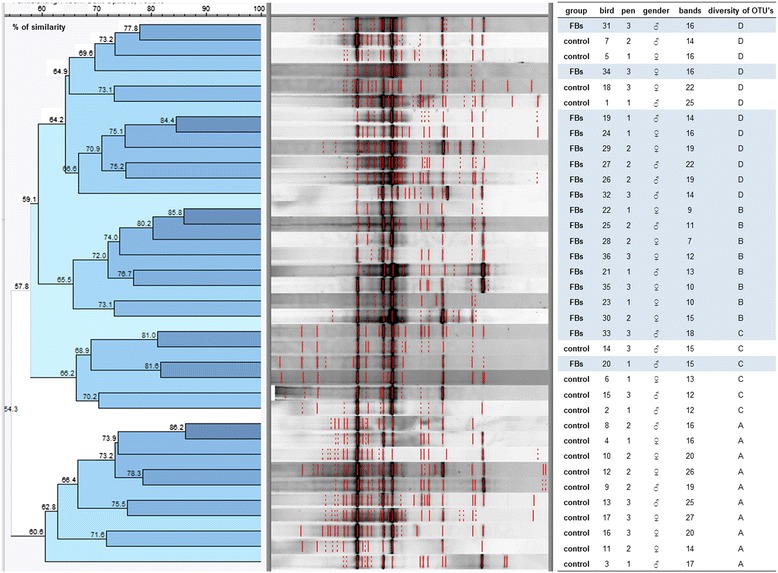
Denaturing gradient gel electrophoresis (DGGE) fingerprint of DNA samples of ileal content applying community PCR with universal bacterial primers targeting the variable V3 region of the 16S ribosomal RNA (18 animals per group (3 pens/group, 6 animals/pen)). Percentage of similarity between DGGE profiles was analyzed using the Dice similarity coefficient, derived from presence or absence of bands. On the basis of a distance matrix, which was generated from the similarity values, dendrograms were constructed using the unweighted pair group method with arithmetic means (UPGMA) as clustering-method. The microbial richness (R) was assessed as the number of OTUs within a profile. Within ileum-samples a separation according to the treatment is visible, (D)although exceptions occur like samples of chicken 20, 31, 33 and 34 of FBs group which show high similarity (69.6 – 81%) with control group. (A)The majority of the control-samples contains bands in the lower GC-range which ascribes them to one clade. (B)Among the FBs group a clade of clearly reduced diversity is formed by 8 samples. (C)The remaining of this treatment group shows a similar diversity of OTUs of medium GC-content like the control-group, however the low-GC-OTUs were absent.
