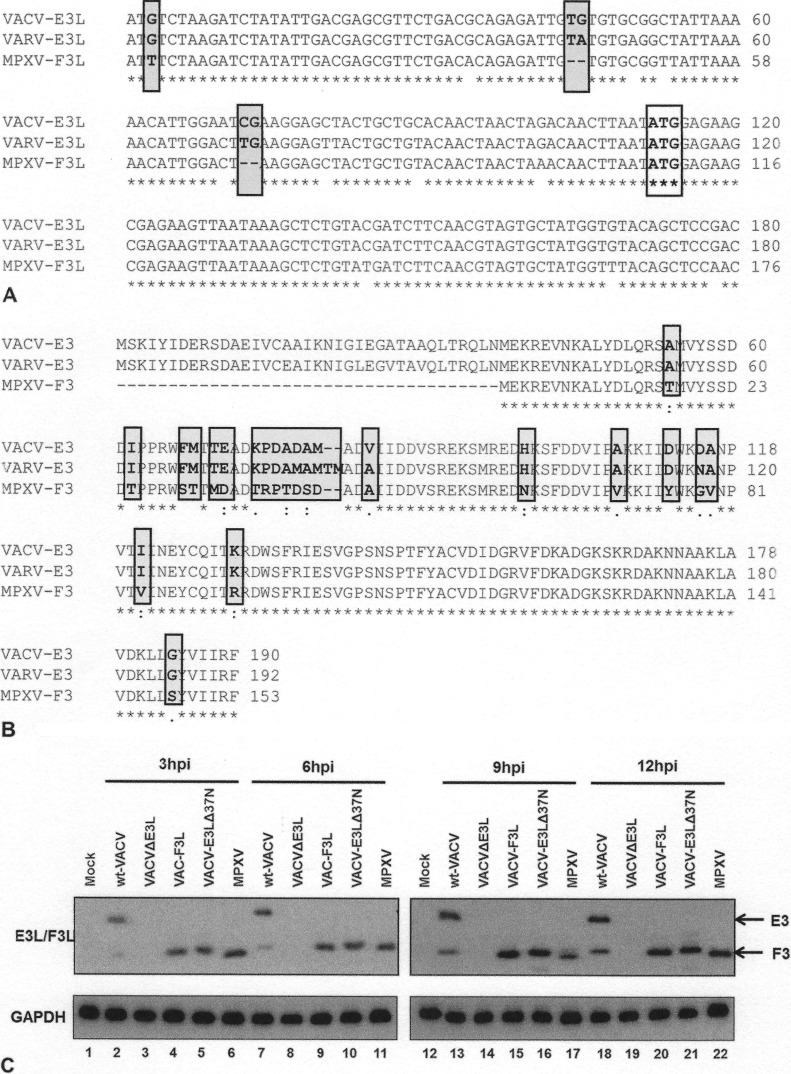
FIG 2.
(A) VACV-E3L homologue alignment. The first 160 nucleotides of the VACV E3L gene homologues of VACV, VARV, and MPXV were aligned by use of the ClustalW program. The sequences begin at nucleotide 1 of each gene, which is the start codon in the case of VACV and VARV. The C-terminal dsRNA BD is highly conserved among all three viruses. Nonhomologous nucleotide differences in MPXV that lead to the inability to generate p25 are shown in gray boxes. The conserved AUG codon that leads to the generation of p20 (the only product expressed by MPXV) is boxed only. Asterisks indicate the nucleotide positions of the consensus sequence among all three sequences. (B) Protein sequence alignment. The VACV E3 protein homologues of VACV, VARV, and MPXV were aligned by use of the ClustalW program. Asterisks, amino acid positions of the consensus sequence among all three sequences; amino acids highlighted in gray, changes in sequences among MPXV, VARV, and VACV; dots, degree of similarity, with one dot being less conservative than two. The N-terminal Z-NA BD for VACV and VARV is highly conserved, whereas the E3 homologue of MPXV (F3) is predicted to contain a 37-amino-acid N-terminal truncation. (C) MPXV produces a p20 form of VACV-E3. HeLa cells were infected with wt VACV, VACV-E3LΔ37N, VACV-F3L, and MPXV at an MOI of 5. At 3 (lanes 2 to 6), 6 (lanes 7 to 11), 9 (lanes 13 to 17), and 12 (lanes 18 to 22) hpi, protein lysates were isolated and analyzed by Western blotting with antibodies specific for the C terminus of E3.