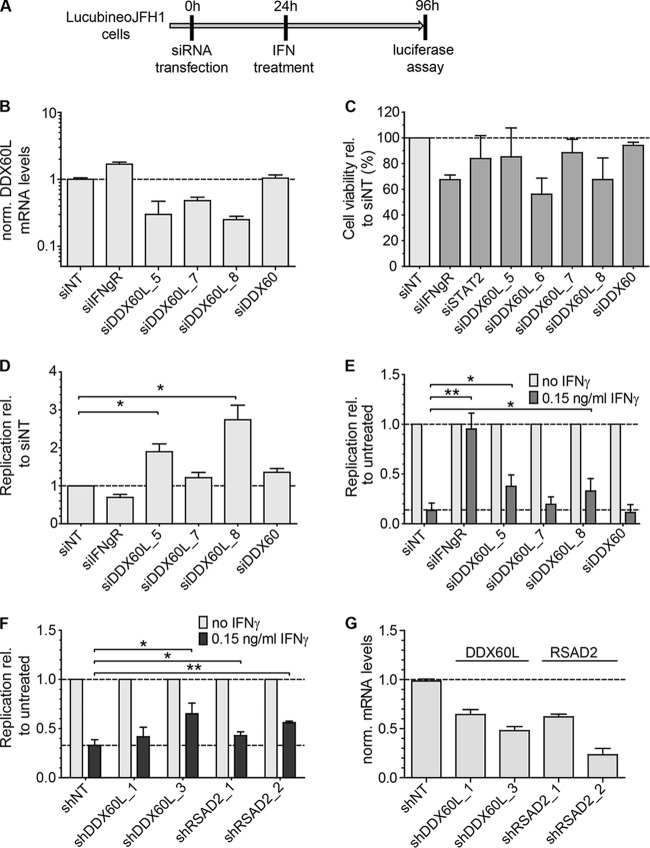
FIG 3.
DDX60L knockdown increases HCV replication in the absence of IFN-γ and partially rescues HCV replication from IFN-γ treatment. (A) Experimental strategy using replicon cell lines with persistent reporter replicons. HCV FLuc reporter replicon cell lines derived from Huh-7-Lunet cells (LucubineoJFH1, genotype 2a) were transfected with siRNAs. Twenty-four hours after transfection, the cells were treated with 0.15 ng/ml of IFN-γ. Ninety-six hours after transfection, HCV replication was determined by measuring luciferase activity. (B) Knockdown efficiency of transfected siRNAs. Forty-eight hours after transfection of siRNAs, DDX60L mRNA levels were determined by qRT-PCR. Shown are mRNA levels relative to that of the siNT control. (C) Cell viability after siRNA transfection. LucubineoJFH1 cells were transfected with siRNAs as for panel B. Ninety-six hours after transfection, cell viability was determined by WST-1 assay and normalized to the siNT control. Shown are mean values with SDs (n = 3). (D) Replication of HCV after siRNA transfection in the absence of IFN-γ. LucubineoJFH1 cells were transfected with indicated siRNAs. Luciferase activity 96 h after transfection was normalized to that of the siNT control. (E) Rescue of HCV replication in the presence of IFN-γ. Light gray bars correspond to bars in panel D, but luciferase activity at 96 h after transfection was normalized to the value for the untreated control of each siRNA, eliminating differences in HCV replication by the siRNA. Given are mean values with SDs (n = 2). (F) Rescue of HCV replication in the presence of IFN-γ using shRNA-mediated knockdown. LucubineoJFH1 cells were transduced with selectable lentiviral vectors encoding shRNAs against DDX60L and RSAD2. After double selection of populations expressing shRNA and replicons, cells were treated with IFN-γ for 72 h and HCV replication was determined by luciferase assay. Results were normalized to those for the untreated control of each shRNA population. Shown are mean values with SDs (n = 4). (G) Knockdown efficiency of shRNAs. Cells were transduced with the corresponding vectors. Ninety-six hours after transduction, DDX6L mRNA levels were determined by qRT-PCR. mRNA levels are shown relative to that of the shNT control.