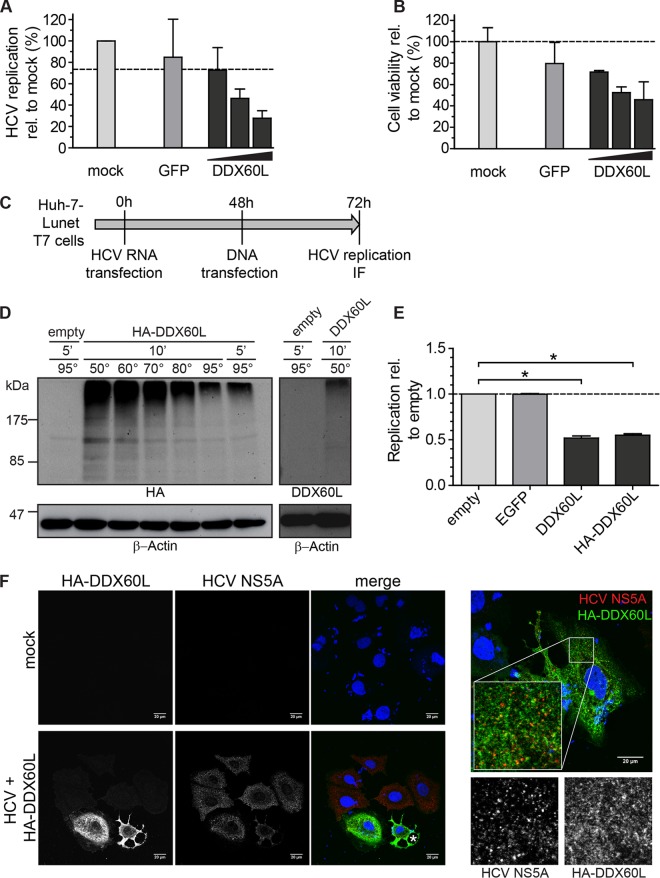
FIG 9.
Ectopic expression of DDX60L using transient transfection. (A and B) DDX60L overexpression under transcriptional control of the CMV promoter by transient transfection of plasmids. LucubineoJFH1 cells were transfected with 300 ng of plasmid, either pcDNA-GFP or 10, 100, or 300 ng of pcDNA-DDX60L, adjusted with pcDNA-GFP to 300 ng or mock transfected, and analyzed 48 h later. Shown are mean values with SDs normalized to values for mock-transfected samples (n = 2). (A) HCV replication was determined by luciferase assay. (B) Cell viability after DDX60L overexpression was determined by WST-1 cell proliferation assay. (C) Experimental strategy for analyzing the impact of DDX60L overexpression on HCV replication using a T7 RNA polymerase-based expression system. Huh-7-Lunet cells expressing T7-RNA polymerase were transfected with HCV FLuc reporter replicons (genotype 2a). After 48 h, the cells were transfected with pTM vectors encoding DDX60L variants, eGFP, or an empty vector control. Twenty-four hours later, cells were harvested and analyzed by luciferase assay for HCV replication (E) or by IF (F). (D) DDX60L protein detection by Western blotting. Protein lysates from Huh-7-Lunet-T7 cells transfected with the indicated pTM constructs were denatured in SDS sample buffer at different temperatures. Denatured lysates were analyzed by SDS-PAGE followed by Western blotting using antibodies detecting HA or DDX60L, as indicated. Note that a lower denaturation temperature led to a slightly better resolution of protein aggregates. β-Actin was used as a loading control. (E) DDX60L overexpression decreases HCV replication. Huh-7-Lunet-T7 cells transfected with HCV Fluc reporter replicon RNA were transfected with pTM vectors encoding DDX60L or eGFP as a control as indicated in panel C. HCV replication was determined by luciferase assay 72 h after transfection of HCV RNA and 24 h after transfection of plasmid DNA. Values are normalized to that of the empty vector control. Shown are mean values with SDs (n = 2). (F) Detection of DDX60L and HCV NS5A by immunofluorescence (IF) microscopy. Cells transfected with HCV replicon RNA and pTM plasmids encoding HA-tagged DDX60L as indicated in panel C or mock-transfected cells were fixed, DDX60L was visualized by IF staining for the HA tag, and HCV replication complexes were detected by using 9E10 antibody directed against NS5A. The HA signal is shown in green, the NS5A signal in red. An asterisk indicates a dying cell.