Fig. 5.
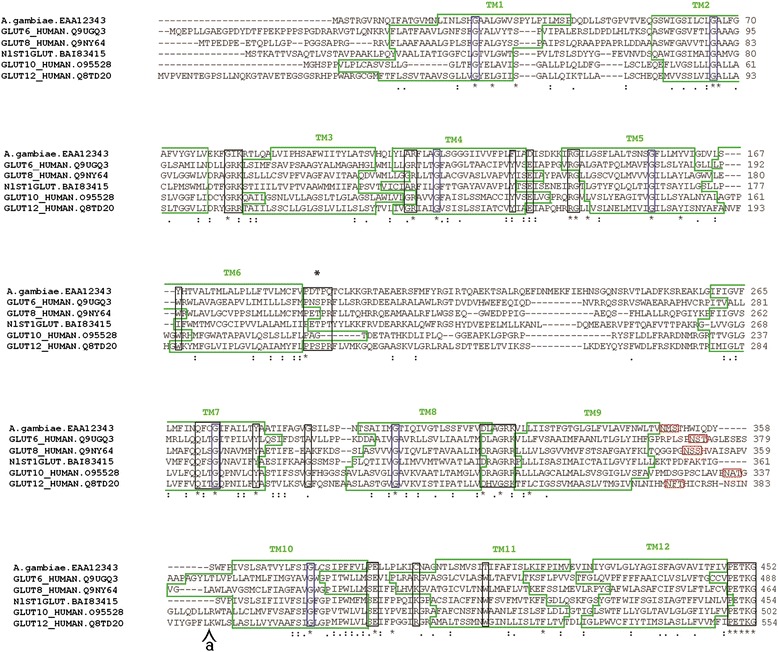
Alignment of members of glucose transporters class III. Class III glucose transporters were aligned using Clustalw. Relevant motifs conserved among class III glucose transporters and An. coluzzii EAA12343 are shown. Annotation of relevant motifs was done using previous reports [95, 96, 98]. Transmembrane domains (TM) are indicated (green boxes), conserved glycine (G) residues (blue boxes), N-linked oligosaccharide site (red boxes) and “PXXPR” motiff (black box with asterisk on top). Other residues and motiff conserved among class III glucose transporters are also shown (black boxes). Insertions of 60 and 68 amino acids in the sequences GLUT10 and GLUT12 respectively were deleted from the alignment, the position is indicate (a). Transmembrane domains were predicted using TMHMM 2.0 [100]
