Abstract
Several enzymes involved in central carbon metabolism and gluconeogenesisplay a critical role in survival and pathogenesis of Mycobacterium tuberculosis (Mtb). The only known functional fructose 1,6-bisphosphatase (FBPase) in Mtb is encoded by the glpX gene and belongs to the Class II sub-family of FBPase. We describe herein the generation of a ΔglpX strain using homologous recombination. Although the growth profile of ΔglpX is comparable to that of wild type Mtb when grown on the standard enrichment media, its growth is dysgonic with individual gluconeogenic substrates such as oleic acid, glycerol and acetate. In mice lung CFU titers of ΔglpX were 2–3 log10 lower than the wild-type Mtb strain. The results indicate that glpX gene encodes a functional FBPase and is essential for both in vitro and in vivo growth and survival of Mtb. Loss of glpX results in significant reduction of FBPase activity but not complete abolition. These findings verify that the glpX encoded FBPase II in Mtb can be a potential target for drug discovery.
Introduction
Mtb grows on a variety of substrates in vitro but mounting evidence indicates that during infection most of its energy is derived from fatty acids [1, 2]. When bacterial metabolism is fueled by fatty acids, synthesis of sugars from intermediates of the TCA cycle (particularly the glyoxylate shunt) become important for growth and persistence [3–6]. Hence, the glyoxylate shunt enzymes malate synthase and isocitrate lyase are considered potential targets for the development of new antibacterial agents [6, 7]. Phosphoenolpyruvate carboxykinase (PEPCK), the enzyme linking the TCA cycle and gluconeogenesis, catalyses the reversible decarboxylation and phosphorylation of oxaloacetate (OAA) to form phosphoenolpyruvate (PEP). The PEPCK-encoding gene pckA is up-regulated by acetate or palmitate, but down-regulated by glucose. Deletion of the pckA gene of Mycobacterium bovis BCG led to a reduction in the capacity of the bacteria to infect and survive in macrophages [8]. PEPCK plays a pivotal role in the pathogenesis of Mtb, as it is essential for growth and survival of this pathogen during infections in mice and Mtb relies primarily on gluconeogenic substrates for in vivo growth and persistence [9]. Except for these recent studies, the role of gluconeogenesis in Mtb pathogenesis has remained largely unaddressed. Therefore, understanding the key structural and functional aspects of enzymes in the gluconeogenic pathway becomes important. Using genetic and biochemical methods, Movahedzadeh et al. have identified Rv1099c gene as a glpX gene that encodes a class II FBPase [10]. Fructose-1,6-bisphosphatase (FBPase, EC 3.1.3.11), a key enzyme of gluconeogenesis, catalyzes the hydrolysis of fructose-1, 6-bisphosphate to form fructose 6-phosphate and orthophosphate. This reaction is the opposite of that catalyzed by phosphofructokinase in glycolysis. Fructose 6-phosphate is the catalytic product of fructose 1,6-bisphosphate, an important precursor in various biosynthetic pathways generating important structural components of the cell wall and glycolipids in mycobacteria. Gluconeogenesis is an important metabolic pathway in several organisms, that allows the cells to synthesize glucose from noncarbohydrate precursors such as organic acids, amino acids, and glycerol. FBPases are members of the large superfamily of lithium-sensitive phosphatases, which includes both the inositol phosphatases and FBPases. These enzymes demonstrate bivalent metal-dependent and lithium-sensitive phosphatase activity [10]. Five different classes of FBPases have been identified based on their amino acid sequences (FBPases I to V) [11–14]. Eukaryotes possess only the FBPase I-type enzyme, but all five types exist in various prokaryotes. Many organisms have more than one FBPase, mostly the combination of types I and II. The type I FBPase is the most widely distributed and is the primary FBPase in Escherichia coli. An additional class II FBPase is encoded by the glpX gene in E. coli, which is part of the glycerol 3-phosphate regulon [11]. The completion of the genome sequence of Mtb allowed the identification of genes that were predicted to encode enzymes for most central metabolic pathways [15]; however, no FBPase was initially assigned. Results from genetic and biochemical analyses revealed that the Rv1099c gene of Mtb encodes the missing mycobacterial FBPase (II) [10]. The protein encoded by the Mtb glpX (Rv1099c) gene is Similar to other class II FBPase from E. coli (GlpX) (42% identity) and Corynebacterium glutamicum FBPase II (65% identity) [16]. The genome wide transposon site hybridization (TraSH) experiment [17] suggests the glpX gene of Mtb to be essential for survival in a mice infection model. Although the method is high throughput and genome wide, it does not characterize individual transposon mutants. To investigate the role of this enzyme in mycobacterial pathogenesis, we generated an unmarked glpX gene deletion knock out (KO) in Mtb and investigated its effect on in vitro and in vivo growth and survival behavior.
Materials and Methods
Cloning and mutagenesis of Mtb glpX gene
Mutagenesis was essentially carried out as previously described [18]. The coding sequence of glpX (Rv1099c) with flanking DNA, 893bp upstream and 970bp downstream of the gene, was amplified by PCR and cloned into p2NIL [18], producing pFM143 (Table 1). The primers used (GlpX_1 and GlpX2) are listed in Table 2. To create a deletion in pFM143, 943bp were deleted in the glpX gene by inverse PCR, using glpX_Rev1 and glpX_Rev2 primers (Table 2)., The PCR product was re-ligated producing FM147. The deletion created was confirmed by sequencing. Following insertion of a gene cassette carrying the lacZ and sacB genes from pGOAL19 into the vector’s PacI site (producing FM152); the DNA was introduced into M. tuberculosis H37Rv by electroporation. Mutagenesis was carried out as previously described [18]. To confirm the mutagenesis, colony PCR and Southern blot were performed.
Table 1. Strains and plasmids used for the generation of Δglpx and glpx complement strains.
| Strains | Characteristics | Source |
|---|---|---|
| M. tuberculosis H37Rv | wild-type laboratory strain | ATCC 27294 |
| HG1 | M. tuberculosis ΔglpX | This study |
| HG2 | M. tuberculosis ΔglpX::pFM163 | This study |
| E.coli DH5α | Invitrogen | |
| p2NIL | manipulation vector | [18] |
| pGOAL19 | delivery cassette vector | [18] |
| pBluescript II SK+ | Stratagene | |
| pUC-Gm-int | [19] | |
| pFM143 | p2NIL::glpx | This study |
| pFM147 | pFM143::glpXΔ | This study |
| pFM152 | pFM147/with PacI cassette of pGoal19 | This study |
| pFM158 | pBluescript SK+::glpX (445/bp upstream) | This study |
| pFM163 | pFM158::intgm | This study |
Table 2. List of primers used for the generation of Δglpx.
| Glpx_Rev1 | AGCGCCGTGTACCCATTGCC |
| Glpx_Rev2 | GTCACCCGGACCAGCTCCAT |
| tbglpX_up | GCTCTGGGTCAAGCTCAGAT |
| tbglpX_end | GGGCAATGGGTACACGGC |
Complementation of ΔglpX
A fragment containing the glpX gene together with 445 bp of upstream sequence was produced by PCR of M. tuberculosis genomic DNA using primers tbglpX_up and tbglpX_end (Table 2), and The PCR product was cloned into the SmaI site of pBluescript-SK+ to produce pFM158. The HindIII cassette of pUC-Gm-int, carrying the int and gm genes was cloned into the HindIII site of pFM158 to produce pFM163. The plasmid was electroporated into HG1, yielding HG2.
Bacterial Strains, media and growth conditions
ΔglpX, glpX complement and WT Mtb H37Rv strains were grown using the shaker flask method in 7H9 liquid medium containing 0.2% glycerol, 10% OADC (Oleic acid, Albumin, Dextrose and Catalase), and 0.05% Tween 80. For growth with defined carbon sources, 7H9 medium was used with 0.05% Tyloxapol and a carbon substrate at 0.1% or 0.2% (wt/vol).
In vivo growth and survival profile of ΔglpX strain in mice model
BALB/c mice (approximately eight weeks old) were used for evaluating the effect of glpX gene knockout on in vivo growth. For each strain, 36 mice were infected via aerosol delivery. At indicated time points 6 mice from each group (infected with the respective strain) were sacrificed via carbon dioxide asphyxiation and the lungs from individual mice were aseptically removed, homogenized and CFU were determined.
Cellular FBPase activity in Mtb strains
Bacteria were grown to mid-log phase, disrupted in a bead beater, the cell-extracts were clarified and FBPase assays were conducted as described previously [20].
Hydrogen peroxide sensitivity
Mtb cultures were grown in 7H9 medium + OADC (100ml) until late log or early stationary phase (Klett units ~ 150) and then treated with 1ml of 500mM hydrogen peroxide solution (effective peroxide concentration = 5mM). 1 ml samples were taken as controls before hydrogen peroxide treatment (referred to as 0 hr /input sample). Additional samples (1 ml) were taken at 2 hrs, 6 hrs and 12 hrs after hydrogen peroxide treatment.
pH sensitivity
Late log or early stationary phase of 100 mL Mtb cultures were treated with 6ml of sterile 2 mM acetic acid solution (effective pH ≈ 4.5). 1 ml sample was taken as control before addition of acetic acid (referred to as 0 hr /input sample). Additional 1ml samples were taken at 2 days, 4 days and 6 days after pH change to ≈ 4.5 units.
Nitrosative stress
Late log or early stationary phase of 100 mL Mtb cultures were treated with 10ml of sterile 2.2 mM DETANO (Diethylenetriamine/nitric oxide) solution (effective DETANO concentration = 0.2 mM). 1ml sample was taken as control before DETANO treatment (referred to as 0 Hr /input sample). Additional 1ml samples were taken at 1 day, 2 days and 3 days after DETANO treatment.
MIC
MIC values for standard drugs were obtained using the standard Microplate-based Alamar Blue Assay (MABA) assay [21].
Results
Unmarked deletion mutant of glpX gene in Mtb
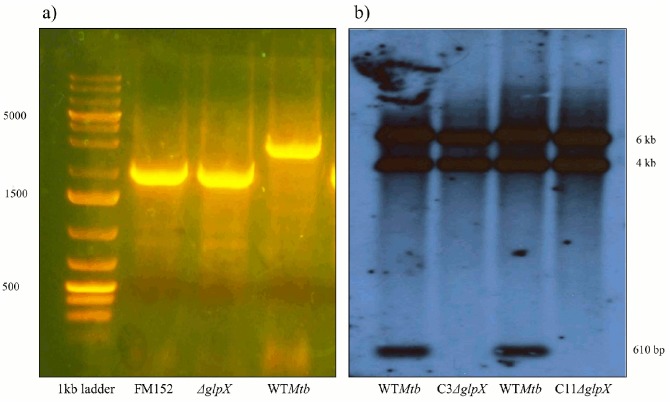
An in-frame deletion of glpX (Rv1099c) was constructed in M. tuberculosis H37Rv producing strain HG1. PCR and Southern blotting analysis were used to confirm the presence of the mutation (Fig 1).
Fig 1.
Colony PCR and Southern Blot confirming the deletion of glpX gene a) Colony PCR results of the potential double cross overs (DCOs): Lane 1: 1kb ladder, Lane 2: PCR amplification product of suicidal delivery vector FM152 is about 2kb, Lane 3: PCR product of one such potential DCO (Colony 11) is 2kb, Lane 4: PCR product of a WT Mtb is about 3kb, b) Southern blot: Lane 1: WT genomic DNA digest with BamH1 which gives a fragment of about 610 bp (lowermost band in lane 1 and 3), Lane 2: Genomic DNA digest of a potential DCO (Colony 3) with the 610 bp fragment missing, Lane 3: Same as Lane 1, Lane 4: Genomic DNA digest of a potential DCO (Colony 11) with the 600 bp fragment missing.
In vitro growth profile of ΔglpX strain (glycerol, dextrose and acetate as carbon sources)
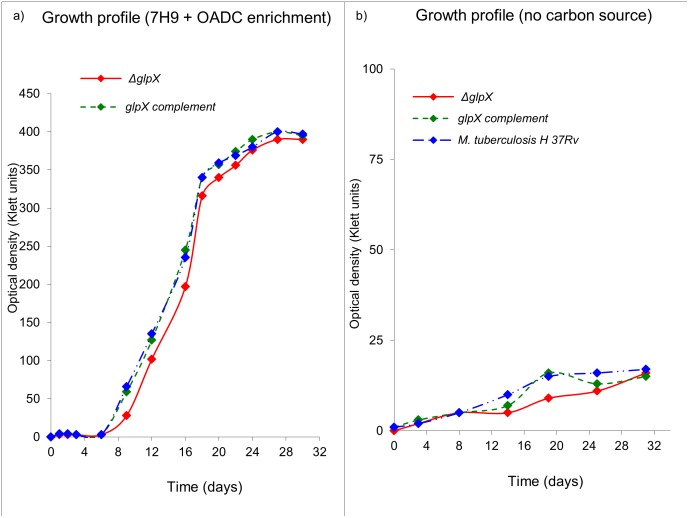
The growth profile of ΔglpX is similar to that of WT Mtb when grown in 7H9 medium with OADC enrichment, plus glycerol and Tween 80, a surfactant which can furnish Mtb with oleic acid (a fatty acid carbon source) through de-esterification [22]. Fig 2 indicates that the disruption of glpX does not significantly affect the growth profile of Mtb, in enriched media (Fig 2a). Both ΔglpX and WT Mtb failed to grow significantly in the absence of any external carbon source (glycolytic/gluconeogenic) as indicated in the growth profile (Fig 2b).
Fig 2. In vitro growth profile of ΔglpX, WT Mtb and glpX complement in a) 7H9 medium + OADC enrichment, b) 7H9 medium with no additional carbon source.
Growth profiles are representative of a triplicate data set.
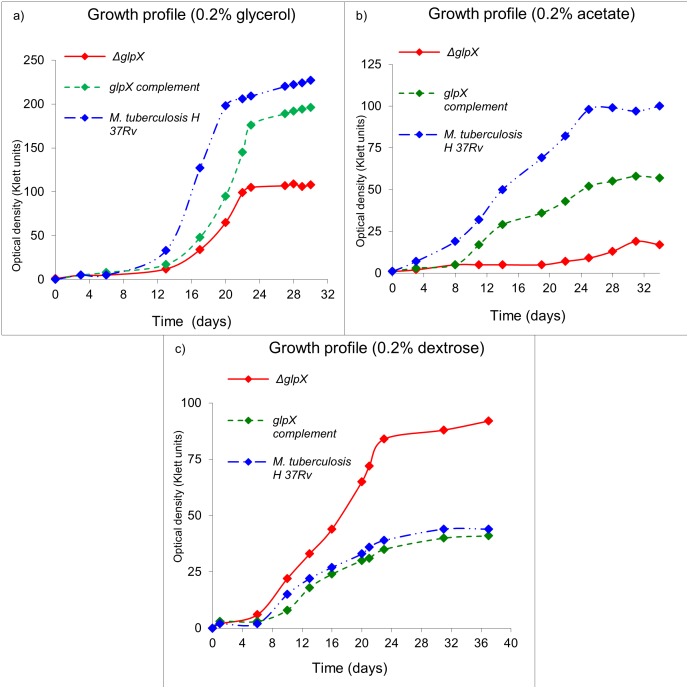
For studies involving growth of ΔglpX on defined carbon sources, defined media was used containing equal concentrations of dextrose, acetate or glycerol as single carbon sources and replacing Tween 80 with a non-hydrolysable detergent, Tyloxapol (Fig 3).
Fig 3. In vitro growth profile of ΔglpX, WT Mtb and glpX complement on defined (or individual) carbon source(s).
Growth profile in 7H9 medium a) 0.2% glycerol, b) 0.2% Acetate, and c) 0.2% dextrose. Growth profiles are representative of a triplicate data set.
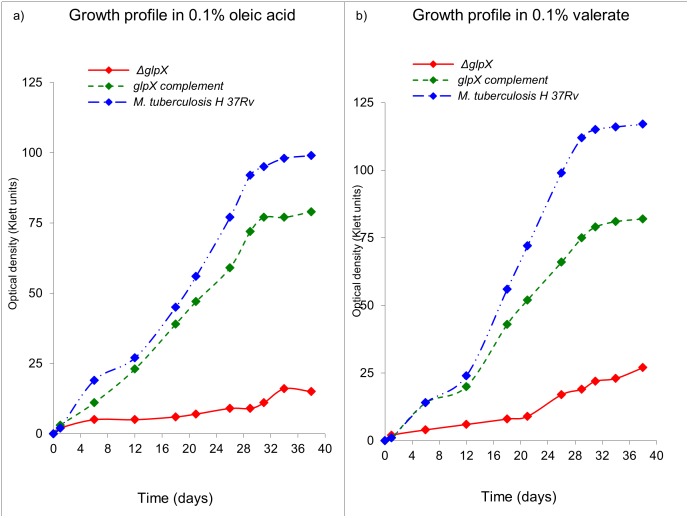
In vitro growth profile of ΔglpX strain on fatty acids as a sole carbon source
As expected and similar to acetate and glycerol based growth profiles, the growth of ΔglpX was severely compromised (Fig 4) on oleic acid (C18) (Fig 4a) or valeric acid (C5) (Fig 4b) as a sole carbon source, further substantiating the observation with glycerol.
Fig 4. In vitro growth profile of ΔglpX, WT Mtb and glpX complement on gluconeogenic carbon source(s).
Growth profile in 7H9 medium a) 0.1% oleic acid, and b) 0.1% valeric acid Growth profiles are representative of a triplicate data set.
In vitro growth profile of ΔglpX strain on combination of carbon sources
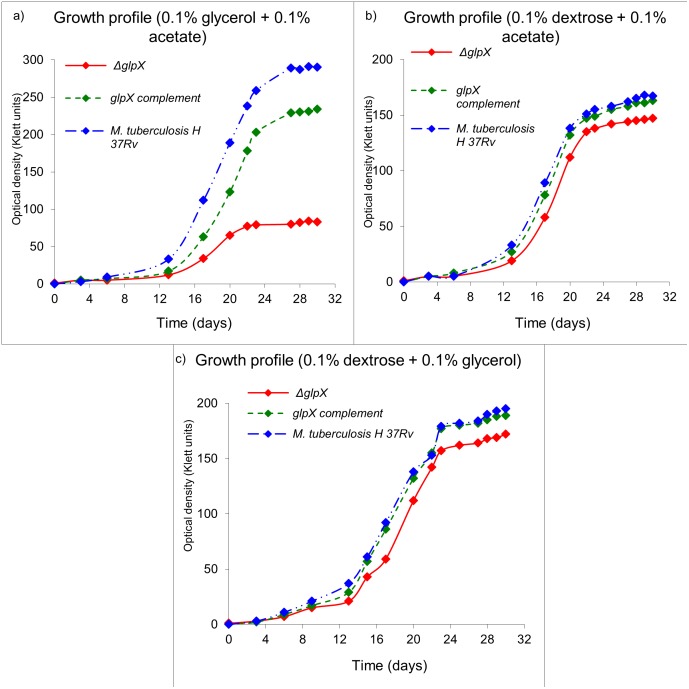
Growth was subsequently monitored using combinations of two carbon sources including acetate, dextrose and glycerol respectively (Fig 5).
Fig 5. In vitro growth profile of ΔglpX, WT Mtb and glpX complement on combination of carbon sources.
Growth profile in 7H9 medium and a) 0.1% each of glycerol and acetate, b) 0.1% each of dextrose and acetate, and c) 0.1% each of dextrose and glycerol. Growth profiles are representative of a triplicate data set.
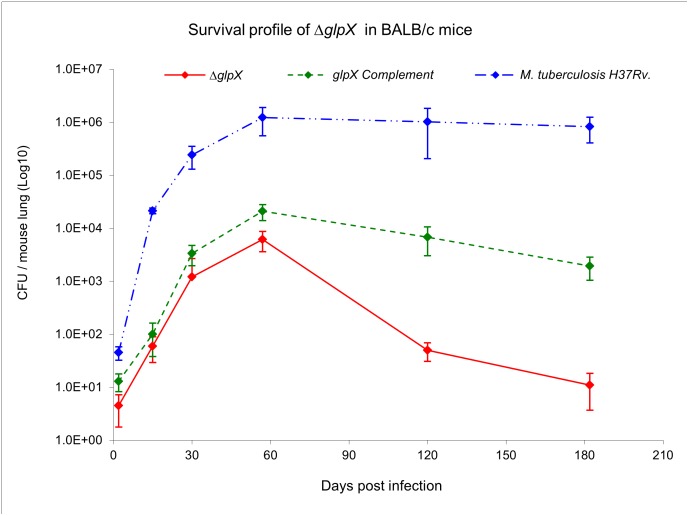
glpX is essential for growth in acute phase and survival during chronic phase of Mtb infection in mice
To determine the role of glpX encoded FBPase in a model of pulmonary tuberculosis, immune-competent BALB/c mice were infected with aerosolized WT Mtb, ΔglpX, and the complemented strains. The aerosolization parameters were set and validated such that they ensured an initial pulmonary bacterial load of about 25–75 CFU/mouse. Such an initial instillation of CFU was achieved for the WT Mtb and the complemented strain but not for the ΔglpX strain, indicating a possible attenuation with regard to infectivity. This difference in initial instillation bacterial count was observed irrespective of the fact that the culture titers of the strains used for infection were all approximately 1 X 106 CFU/ml (data not shown). It is clear that the ΔglpX strain failed to efficiently replicate during the acute phase (i.e. first 30 days post-infection) of infection as compared to WT Mtb. The CFU for the ΔglpX strain were consistently 2–3 log10 lower than those for WT Mtb. The CFU for ΔglpX in lungs started to decline rapidly after day 57 (Fig 6) and continued to remain low 120 and 180 days post-infection. Introduction of glpX expressed from its native promoter (glpX complement strain) restored survival and replication through both acute and chronic phases, although the bacterial load in the lungs did not reach the WT level (data not shown). Similar to the in vitro growth profile of the glpX complemented strain on fatty acids or gluconeogenic growth substrates, growth in vivo was not fully restored to WT levels (Fig 6) Such a behavior has been observed for complemented strains of several mutants [9, 23, 24]. It is noteworthy that a glpX gene deletion does not completely abolish FBPase activity in Mtb (Fig 7). These experiments demonstrate that glpX is not only essential for Mtb to establish an infection and to grow during the acute phase of infection, but is equally important for survival of Mtb during the chronic phase of infection.
Fig 6. glpX is essential for growth in acute phase and survival during the chronic phase of Mtb infection in mice.
Invivo growth and survival plot for ΔglpX compared to WT Mtb and the glpX complement. Data represents the mean±s.d. of six mice per time point.
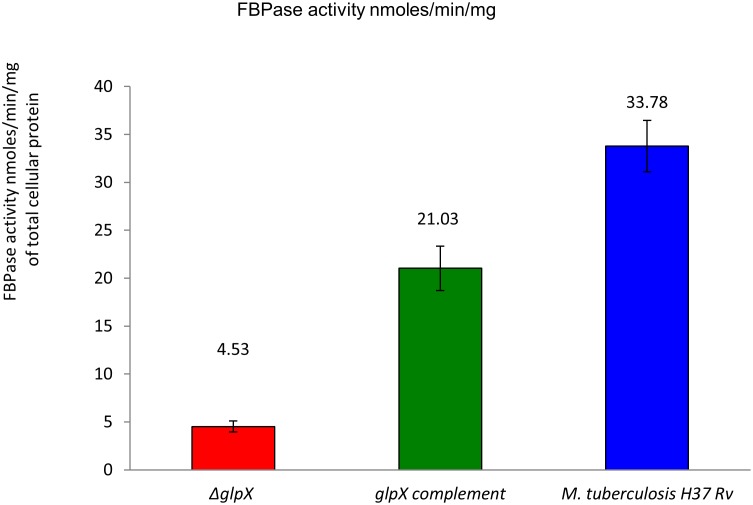
Fig 7. glpX gene deletion does not completely abolish the FBPase activity in Δglpx: FBPase Activity was measured in nmol/min/mg protein in crude extracts, mean of two determinations, limit of detection = 0.4.
All values are with a substrate-free control (no FBP) subtracted. The reported readings are the average of 6 measurements coming from duplicate protein samples (n = 3X2 = 6).
Fructose 1,6-bisphosphatase activity in cell extracts
The total cellular FBPase activity in WT, ΔglpX and complemented strains indicated that the deletion of glpX gene does not completely abolish FBPase activity (ΔglpX activity = 4.53 ± 0.57 nmol/min/mg), and while the glpX complement strain restored the FBPase activity levels (FBPase activity = 21.03 ± 2.31 nmol/min/mg), it does not restore it to the WT levels (FBPase activity = 33.78 ± 2.67 nmol/min/mg). 2 mM of F1,6BP was used as substrate in this assay.
ΔglpX sensitivity to stress factors
The survival of the knockout under various in vitro stress conditions (low pH, hydrogen peroxide and nitric oxide) was measured and no significant changes were observed in comparison with Wild type (data not shown).
MIC for ΔglpX strain is comparable to that for WT Mtb for all the standard drugs tested except for PA-824
The inhibitory concentrations for both strains were not strikingly different except for PA-824 (Table 3).
Table 3. MICs for standard antibacterial drugs as determined by the MABA assay.
| MIC (μM) a | ||
|---|---|---|
| Standard drugs | ΔglpX | WT Mtb H37Rv |
| RMP | 0.023 ± 0.005 | 0.053 ± 0.007 |
| INH | 0.479 ± 0.010 | 0.476 ± 0.009 |
| MET | > 512 | > 512 |
| CAP | 1.72 ± 0.061 | 1.89 ± 0.095 |
| SM | 0.48 ± 0.097 | 0.82 ± 0.083 |
| PA-824 | 0.48 ± 0.027 | 0.16 ± 0.007 |
| OPC-67683 | 0.006 ± 0.001 | 0.009 ± 0.001 |
a Reported MIC values are an average (± standard deviation) of 6 independent assays. Although the MIC of PA-824 for the ΔglpX strain is significantly higher (about 3–4 fold) than that observed for WT Mtb strain, it is within the normal MIC range of 0.015 to 0.5 μM.
Discussion
As expected, in aerated batch cultures, WT Mtb strain grew fastest and achieved the highest growth rate on glycerol, followed by dextrose and acetate, similar to the earlier studies [25]. While the ability of ΔglpX to grow on glycerol or acetate as a sole carbon source was severely compromised as compared to WT Mtb, ΔglpX grew twice as fast as WT Mtb on dextrose as a sole carbon source. The slower growth profile of ΔglpX on glycerol or acetate was expected since glpX encodes a functional fructose 1,6-bisphosphatase (FBPase) required for growth on gluconeogenic substrates (glycerol/fatty acids/acetate) [10, 20]. Previous studies [4, 26–30] indicate that Mtb grows fastest in vitro on glycerol, less quickly on dextrose, and least quickly on acetate.
The fast growth of ΔglpX on dextrose is most likely a result of the disruption of a possible regulatory mechanism of FBPase with the phosphofructokinase (encoded by pfk1 and pfk2 in Mtb) enzyme (which catalyzes the reverse reaction converting fructose 6-phosphate to fructose 1,6-bisphosphate). However the presence of such a regulatory mechanism for FBPase is not experimentally verified in Mtb. Taken together the observations indicate the ΔglpX does not display an eugonic growth phenotype on gluconeogenic substrates (acetate, glycerol, fatty acids), rather it is dysgonic. This observation is concordant with the expected disruption of FBPase activity in Mtb by deletion of the glpX gene. The growth of WT Mtb on an equimolar mixture of glycerol and acetate exceeded that achieved with either constituent alone. Similar effects were observed using a mixture of dextrose and glycerol and a mixture of acetate and dextrose. The compromised growth of ΔglpX on glycerol or acetate was not completely rescued by their combination either. However, with the presence of dextrose in the medium (dextrose + glycerol or dextrose + acetate), the growth profile was almost similar to WT Mtb. A reason for such a phenotype is that both glycerol and acetate are gluconeogenic in nature and therefore cannot be effectively utilized by the ΔglpX. However, dextrose being a preferred glycolytic nutrient for ΔglpX easily rescues the glpX mutant strain and the combinations involving dextrose grew in a manner similar to WT Mtb. Central carbon metabolism in Mtb is a peculiar case involving compartmentalization of metabolic pathways and thereby the metabolites. As understood from our results and the model of compartmentalized central carbon metabolism [25], Mtb preferentially utilizes glycerol over dextrose and acetate. The predominant pattern of distribution for glycerol and acetate combination as carbon source requires a functional glpX/FBPase. However, since ΔglpX does not possess a fully functional FBPase, its growth is severely compromised on gluconeogenic substrates like glycerol and acetate. The reduced growth/growth-compromised phenotype of ΔglpX is rescued to some extent by dextrose in combination growth media since with dextrose as a carbon source, FBPase activity becomes dispensable (predominant pattern of distribution is not gluconeogenic but rather glycolytic).
As anticipated, glpX gene deletion adversely affected the FBPase activity levels in ΔglpX. However, FBPase activity was not completely abolished in the ΔglpX indicating a possible compensation by other genes/alternate proteins having FBPase activity. The residual FBPase activity in ΔglpX can be attributed to several other gene products/ other classes of enzymes such as inositol monophosphatases (IMPase). The M. tuberculosis genome encodes four IMPase like genes, ImpA, SuhB (Rv2701c), ImpC (Rv3137) and CysQ (Rv2131c) [15, 31, 32]. The SuhB of Mtb has been extensively characterized both structurally and biochemically [31, 33]. SuhB appears to be a bona fide IMPase with no activity towards fructose-1,6-bisphosphate. The purified CysQ (Rv2131c) gene product, in addition to having IMPase and FBPase activity, showed substrate specificity that was broader than those of several bacterial and eukaryotic IMPase. The dimeric enzyme exhibited dual activities of IMPase and FBPase, with Km of 0.22 ± 0.03 mM for IMP and Km of 0.45 ± 0.05 mM for FBP. All four IMPases in Mtb share very limited sequence homology with each other (<30% identity with the best being SuhB which is 30% identical to ImpC (Rv3137)). Both ImpA and ImpC share similar sequence homology (20–25% sequence identity) with TM1415 IMPase/FBPase–Thermotoga maritime (PDB id: 2P3N) [34] and MJ0109 IMPase/FBPase–Methanococcus jannaschii (PDB id: 1G0H) [35]. It is also noteworthy that Both ImpA and ImpC have not been characterized structurally or biochemically yet to rule out the possibility of a dual function IMPase/FBPase. Taken together the residual FBPase activity in ΔglpX is attributable to CysQ or ImpA and ImpC.
The in vivo survival profile suggests that glpX is required to achieve an initial instillation dose and also maintain high bacterial loads at later time points (acute and chronic phases). The initial instillation dose for ΔglpX is significantly lower than the WT Mtb and complement respectively. There are several reports wherein the initial bacterial loads of certain gene deletion strains of Mtb are significantly different than the control WT Mtb strain [24, 36, 37]. In the study by Venugopal et al., in describing the phenotype of ΔdlaTΔpdhC, the deletion strain was not recovered post-day one despite equal input into the aerosolizer. However, by day 7, the ΔdlaTΔpdhC and ΔlpdC mutants were both recovered at 100 CFU, suggesting that the ΔdlaTΔpdhC mutant was viable but non-culturable at day one. We expect a similar case for ΔglpX, where the deletion of glpX has limited the viability of the bacteria. The lower bacterial counts at later time points could be due to an initially low instillation dose (<100 CFU at 1 day post infection), however the rate of growth differs during the initial 30 days post-infection. The significance or essentiality of genes can be efficiently studied at varying stages of disease progression (acute vs. chronic infection) by generating conditional mutants to understand the function of such essential genes. ΔglpX strain like WT Mtb, is sensitive to stress factors likely to be encountered inside the host, such as low pH, hydrogen peroxide, and nitric oxide indicating that the growth suppressed phenotype in vivo is primarily due to a drastic reduction of FBPase activity and not to any other underlying effects.
The ΔglpX strain showed similar MICs for all the drugs tested except for PA-824. The glpX gene deletion reduces the inhibitory activity of PA-824 but not OPC-67683, a structurally similar nitroimidazole. Rv3547, encoding a 151 amino acid protein with no similarity to any protein with a known function, was characterized as a F420-dependent nitroreductase [38, 39]. F420-dependent glucose-6-phosphate dehydrogenase, which catalyzes the oxidation of G6P to 6-phosphogluconolactone, is required for the intracellular reduction of the deazaflavin cofactor F420, which serves as the hydride donor to PA-824 in the Rv3547 catalyzed reduction of this compound. Mutations in the mycobacterial Rv3547 gene are found in OPC-67683-resistant Mtb strains as well, suggesting that the Rv3547-encoded enzyme is required for activation of both PA-824 and OPC-67683 [40]. However, there is no conclusive evidence whether FGD1 and coenzyme F420 are also needed for activation of OPC-67683, since there were no OPC-67683 resistant strains with variations (mutations) in FGD1 or coenzyme F420. Therefore it is possible that the activation mechanism of OPC-67683 is FGD1/F420 independent which would be consistent with our results wherein the MIC of OPC-67683 for ΔglpX strain is comparable to that of WT Mtb strain. The reduced FBPase activity in ΔglpX strains could result in lower intracellular F6P/G6P levels and hence reduced G6P dehydrogenase activity and the corresponding reduced hydride donor activity of F420. Thus the modest resistance of ΔglpX to PA-824 may be due to reduced intracellular G6P/F6P levels.
In conclusion the glpX gene encodes a functional FBPase and is essential for both in vitro and in vivo growth and survival of Mtb. The loss of this gene results in drastic reduction of FBPase activity but not complete abolition. Our findings verify the glpX encoded FBPase II in Mtb can be a potential target for drug discovery.
Acknowledgments
We would like to acknowledge Dr Neil Stoker since the KO constructs were constructed in his laboratory at RVC, London, UK and Jasper Marc Bondoc for assistance with proof reading of the manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the American Lung Association (Grant No. RG-82534-N), and the Chicago Biomedical Consortium with support from The Searle Funds at the Chicago Community Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol. 2009;10(9):943–8. 10.1038/ni.1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boshoff HI, Barry CE 3rd. Tuberculosis—metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3(1):70–80. . [DOI] [PubMed] [Google Scholar]
- 3. McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406(6797):735–8. . [DOI] [PubMed] [Google Scholar]
- 4. Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11(6):638–44. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dunn MF, Ramirez-Trujillo JA, Hernandez-Lucas I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology. 2009;155(Pt 10):3166–75. 10.1099/mic.0.030858-0 [DOI] [PubMed] [Google Scholar]
- 6. Sharma V, Sharma S, Hoener zu Bentrup K, McKinney JD, Russell DG, Jacobs WR Jr, et al. Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat Struct Biol. 2000;7(8):663–8. . [DOI] [PubMed] [Google Scholar]
- 7. Smith CV, Huang CC, Miczak A, Russell DG, Sacchettini JC, Honer zu Bentrup K. Biochemical and structural studies of malate synthase from Mycobacterium tuberculosis. J Biol Chem. 2003;278(3):1735–43. . [DOI] [PubMed] [Google Scholar]
- 8. Liu K, Yu J, Russell DG. pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and in macrophages. Microbiology. 2003;149(Pt 7):1829–35. . [DOI] [PubMed] [Google Scholar]
- 9. Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci U S A. 2010;107(21):9819–24. 10.1073/pnas.1000715107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Movahedzadeh F, Rison SC, Wheeler PR, Kendall SL, Larson TJ, Stoker NG. The Mycobacterium tuberculosis Rv1099c gene encodes a GlpX-like class II fructose 1,6-bisphosphatase. Microbiology. 2004;150(Pt 10):3499–505. . [DOI] [PubMed] [Google Scholar]
- 11. Donahue JL, Bownas JL, Niehaus WG, Larson TJ. Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol 3-phosphate regulon of Escherichia coli. J Bacteriol. 2000;182(19):5624–7. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nishimasu H, Fushinobu S, Shoun H, Wakagi T. The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea. Structure. 2004;12(6):949–59. . [DOI] [PubMed] [Google Scholar]
- 13. Hines JK, Chen X, Nix JC, Fromm HJ, Honzatko RB. Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition. J Biol Chem. 2007;282(49):36121–31. . [DOI] [PubMed] [Google Scholar]
- 14. Hines JK, Fromm HJ, Honzatko RB. Novel allosteric activation site in Escherichia coli fructose-1,6-bisphosphatase. J Biol Chem. 2006;281(27):18386–93. . [DOI] [PubMed] [Google Scholar]
- 15. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–44. . [DOI] [PubMed] [Google Scholar]
- 16. Rittmann D, Schaffer S, Wendisch VF, Sahm H. Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and biochemical characterization of the enzyme. Arch Microbiol. 2003;180(4):285–92. . [DOI] [PubMed] [Google Scholar]
- 17. Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48(1):77–84. Epub 2003/03/27. 3425 [pii]. . [DOI] [PubMed] [Google Scholar]
- 18. Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146 (Pt 8):1969–75. Epub 2000/08/10. . [DOI] [PubMed] [Google Scholar]
- 19. Mahenthiralingam E, Marklund BI, Brooks LA, Smith DA, Bancroft GJ, Stokes RW. Site-directed mutagenesis of the 19-kilodalton lipoprotein antigen reveals No essential role for the protein in the growth and virulence of Mycobacterium intracellulare. Infect Immun. 1998;66(8):3626–34. Epub 1998/07/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gutka HJ, Rukseree K, Wheeler PR, Franzblau SG, Movahedzadeh F. glpX gene of Mycobacterium tuberculosis: heterologous expression, purification, and enzymatic characterization of the encoded fructose 1,6-bisphosphatase II. Appl Biochem Biotechnol. 2011;164(8):1376–89. Epub 2011/04/01. 10.1007/s12010-011-9219-x . [DOI] [PubMed] [Google Scholar]
- 21. Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, et al. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol. 1998;36(2):362–6. Epub 1998/02/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dubos RJ, Davis BD, et al. The effect of water soluble lipids on the growth and biological properties of tubercle bacilli. Am Rev Tuberc. 1946;54(3):204–12. Epub 1946/09/01. . [DOI] [PubMed] [Google Scholar]
- 23. Movahedzadeh F, Smith DA, Norman RA, Dinadayala P, Murray-Rust J, Russell DG, et al. The Mycobacterium tuberculosis ino1 gene is essential for growth and virulence. Mol Microbiol. 2004;51(4):1003–14. Epub 2004/02/07. . [DOI] [PubMed] [Google Scholar]
- 24. Awasthy D, Ambady A, Bhat J, Sheikh G, Ravishankar S, Subbulakshmi V, et al. Essentiality and functional analysis of type I and type III pantothenate kinases of Mycobacterium tuberculosis. Microbiology. 2010;156(Pt 9):2691–701. Epub 2010/06/26. mic.0.040717-0 [pii]. [DOI] [PubMed] [Google Scholar]
- 25. de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol. 2010;17(10):1122–31. Epub 2010/11/03. S1074-5521(10)00347-9 [pii] 10.1016/j.chembiol.2010.08.009 . [DOI] [PubMed] [Google Scholar]
- 26. Youmans AS, Youmans GP. Studies on the metabolism of Mycobacterium tuberculosis. IV. The effect of fatty acids on the growth of M. tuberculosis var. hominis. J Bacteriol. 1954;67(6):731–3. Epub 1954/06/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Youmans GP, Youmans AS. Studies on the metabolism of Mycobacterium tuberculosis. I. The effect of carbohydrates and alcohols on the growth of Mycobacterium tuberculosis var. hominis. J Bacteriol. 1953;65(1):92–5. Epub 1953/01/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Youmans AS, Youmans GP. Studies on the metabolism of Mycobacterium tuberculosis. II. The effect of compounds related to the Kreb's tricarboxylic acid cycle on the growth of Mycobacterium tuberculosis var. hominis. J Bacteriol. 1953;65(1):96–9. Epub 1953/01/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Youmans AS, Youmans GP. Studies on the metabolism of Mycobacterium tuberculosis. III. The growth of Mycobacterium tuberculosis var. hominis in the presence of various intermediates of the dissimilation of glucose to pyruvic acid. J Bacteriol. 1953;65(1):100–2. Epub 1953/01/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Youmans AS, Youmans GP. Studies on the metabolism of Mycobacterium tuberculosis. V. The effect of amino acids on the growth of M. tuberculosis var. hominis. J Bacteriol. 1954;67(6):734–7. Epub 1954/06/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nigou J, Dover LG, Besra GS. Purification and biochemical characterization of Mycobacterium tuberculosis SuhB, an inositol monophosphatase involved in inositol biosynthesis. Biochemistry. 2002;41(13):4392–8. . [DOI] [PubMed] [Google Scholar]
- 32. Movahedzadeh F, Wheeler PR, Dinadayala P, Av-Gay Y, Parish T, Daffe M, et al. Inositol monophosphate phosphatase genes of Mycobacterium tuberculosis. BMC Microbiol. 2010;10:50 Epub 2010/02/20. 1471-2180-10-50 [pii] 10.1186/1471-2180-10-50 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown AK, Meng G, Ghadbane H, Scott DJ, Dover LG, Nigou J, et al. Dimerization of inositol monophosphatase Mycobacterium tuberculosis SuhB is not constitutive, but induced by binding of the activator Mg2+. BMC Struct Biol. 2007;7:55 Epub 2007/08/30. 10.1186/1472-6807-7-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen L, Roberts MF. Characterization of a tetrameric inositol monophosphatase from the hyperthermophilic bacterium Thermotoga maritima. Appl Environ Microbiol. 1999;65(10):4559–67. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen L, Roberts MF. Cloning and expression of the inositol monophosphatase gene from Methanococcus jannaschii and characterization of the enzyme. Appl Environ Microbiol. 1998;64(7):2609–15. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Venugopal A, Bryk R, Shi S, Rhee K, Rath P, Schnappinger D, et al. Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe. 2011;9(1):21–31. Epub 2011/01/18. S1931-3128(10)00414-2 [pii] 10.1016/j.chom.2010.12.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun R, Converse PJ, Ko C, Tyagi S, Morrison NE, Bishai WR. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol Microbiol. 2004;52(1):25–38. Epub 2004/03/31. 10.1111/j.1365-2958.2003.03958.x MMI3958 [pii]. . [DOI] [PubMed] [Google Scholar]
- 38. Singh R, Manjunatha U, Boshoff HI, Ha YH, Niyomrattanakit P, Ledwidge R, et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322(5906):1392–5. Epub 2008/11/29. 322/5906/1392 [pii] 10.1126/science.1164571 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manjunatha UH, Lahiri R, Randhawa B, Dowd CS, Krahenbuhl JL, Barry CE 3rd. Mycobacterium leprae is naturally resistant to PA-824. Antimicrob Agents Chemother. 2006;50(10):3350–4. Epub 2006/09/29. 50/10/3350 [pii] 10.1128/AAC.00488-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, et al. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 2006;3(11):e466 Epub 2006/11/30. 06-PLME-RA-0146R3 [pii] 10.1371/journal.pmed.0030466 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.