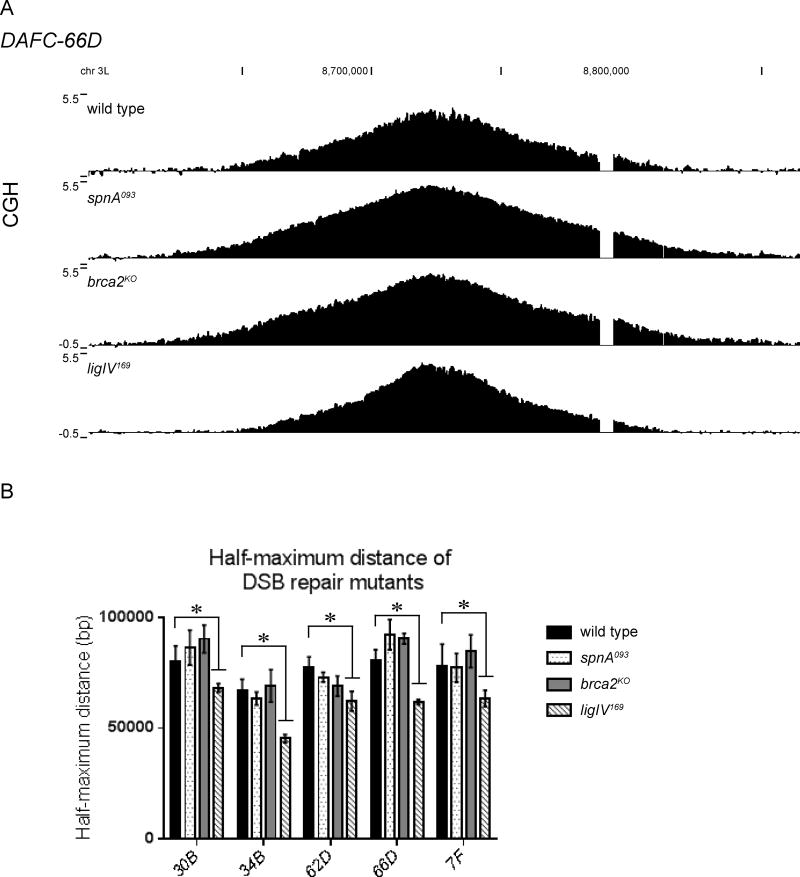
Figure 4. LigIV is utilized for DSB repair during re-replication.
(A) CGH of DAFC-66D reveals impaired replication fork progression in the ligIV169, but not the spnA093 or brca2KO mutants. DNA from stage 13 egg chambers was competitively hybridized with diploid embryonic DNA to microarrays with approximately one probe every 125bp. Chromosomal position is plotted on the x-axis, the log2 ratio of stage 13 DNA to embryonic DNA is plotted on the y-axis.
(B) The half-maximum distance was calculated in the wild-type and mutant backgrounds for each DAFC. Each half-maximum value is the average of three biological replicates. Significance measured by the Dunnett test for multiple comparisons, asterisks indicate p<0.05. The spnA093 and brca2KO mutants are not significantly different from wild type.