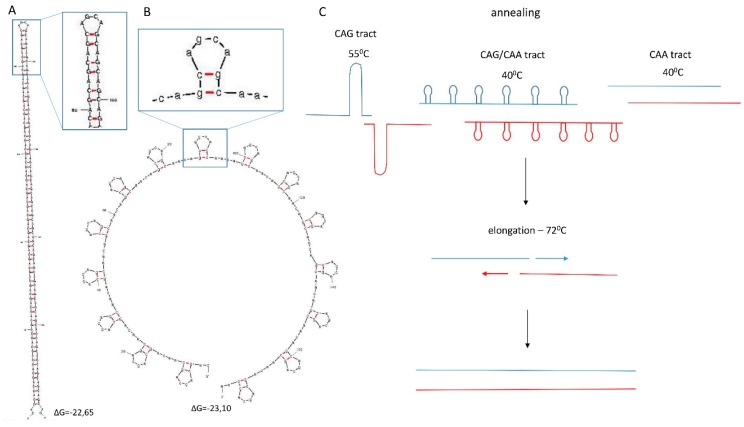
Figure 4.
Structure of CAG and (CAG)3CAA repeat tracts predicted for 60 triplets using Mfold (DNA folding form), and hypothetical hairpin structure behavior during SLIP. Red and blue colors indicate products of BsmBI and Eco0109I restriction enzymes digestions. Predictions were performed at default settings using an annealing temperature corresponding to the temperature used during the PCR cycle. (A) CAG repeat structure at 55 °C; (B) (CAG)3CAA repeat structure at 40 °C. Only the lowest energy structures are presented; (C) hypothetical behavior of repeat tracts during SLIP. At annealing temperature, hairpin structures are relatively stable, with ΔG = −22.65 kcal/mol for the CAG tract and ΔG = −23.15 kcal/mol for the CAG/CAA tract; at 72 °C, the ΔG rises to −6.1 and 0.62, respectively, making hairpins less stable and allowing polymerase to function [32,33,34].