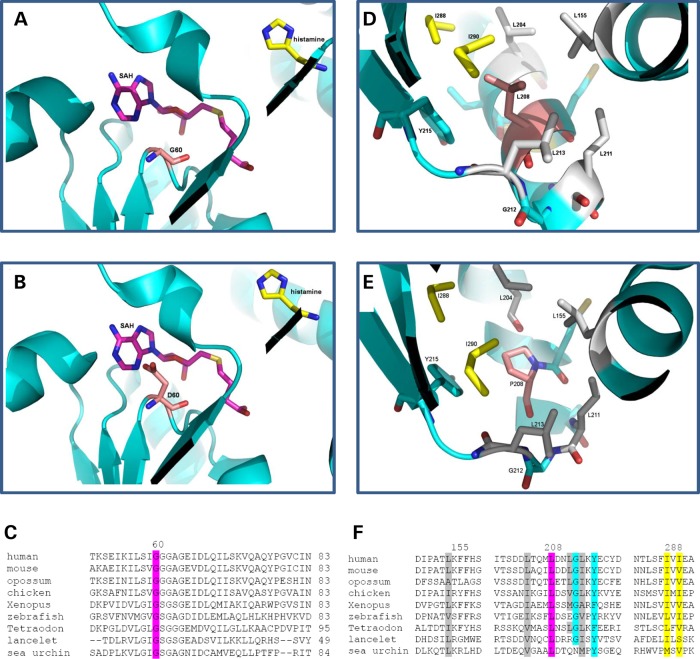
Figure 3.
Three-dimensional Protein structure and ClustalW2 analysis: structures of HNMT (pdb 1jqd) at the catalytic domain for (A) Gly60, (B) Asp60, with HA in yellow and S-adenosyl homocysteine (SAH) in pink, and Gly60 and Asp60 in light pink, and (C) ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2) alignment/comparison of HNMT across vertebrate species showing conservation at Gly60Asp (highlighted in pink). Structures at the hydrophobic pocket around residue 208 (labeled pink) for (D) Leu208 and (E) Pro208. Leu155, Leu204, Leu211 and Leu213 were labeled in gray. Ile288 and Ile290 were labeled in yellow. Tyr215 and Gly212 were labeled in blue. (F) ClustalW2 alignment of HNMT for the hydrophobic pocket surrounding Leu208, with Leu208 highlighted in pink, Leu155, Leu204, Leu211 and Leu213 labeled in gray, Ile288 and Ile290 labeled in yellow. Tyr215 and Gly212 labeled in blue. Sequences used for the HNMT alignment included human (NP_008826.1), mouse (NP_536710.1), opossum (from N-SCAN and Genscan gene predictions, UCSC browser), chicken (NP_001264802.1), Xenopus laevis (NP_001080614.1), zebrafish (NP_001003636.1), Tetraodon nigroviridis (Q4SBY6.1), lancelet (Branchiostoma floridae: predicted from mRNA XM_002613293.1) and sea urchin (Strongylocentrotus purpuratus: from mRNAs CX698504, CD312314 and CX689147).