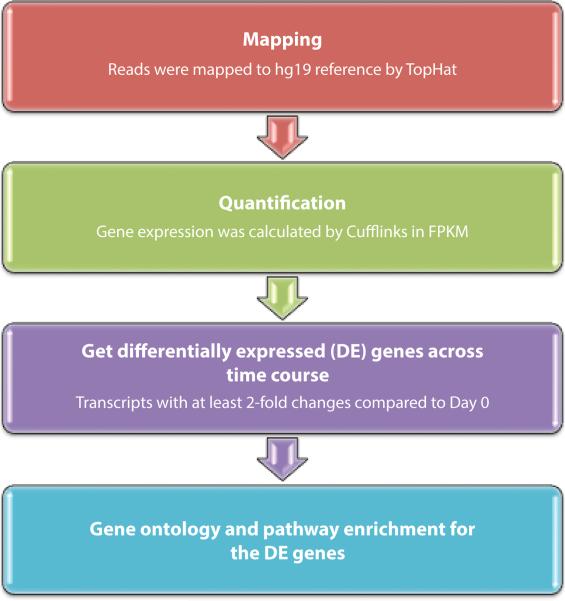
Figure 2.
Schematic of RNA-Seq data processing. RNA-Seq reads were mapped to the hg19 reference genome using TopHat. Gene expression was calculated based on FPKM using Cufflinks. Greater than two-fold change in expression was used to define differentially expressed (DE) loci for subsequent evaluation. Finally, Gene Ontology term analysis was performed for differentially expressed genes.