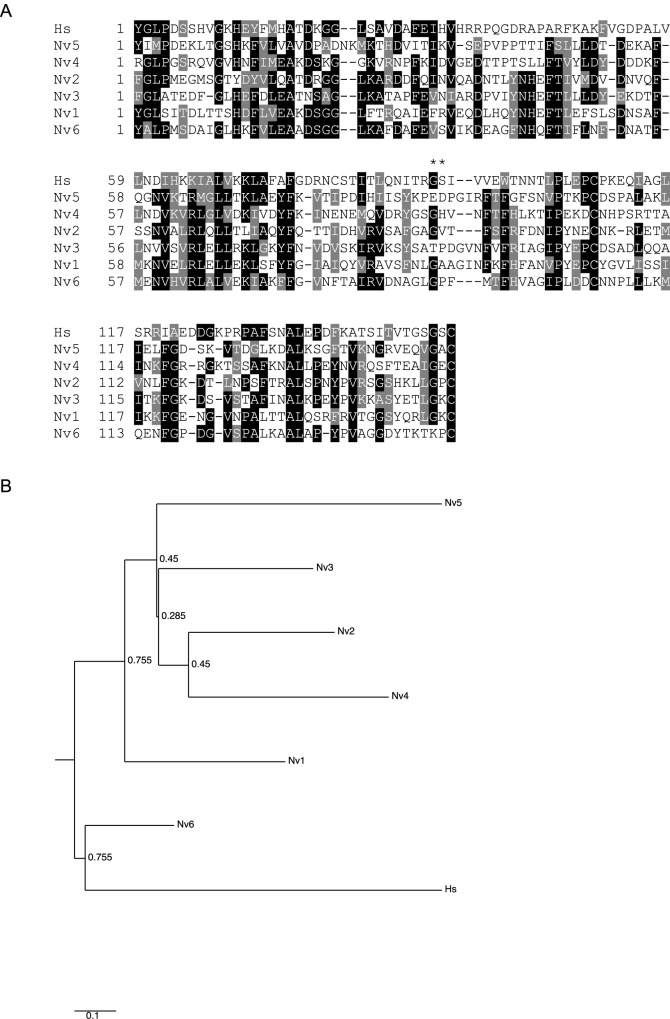
Fig. 2.
. Analysis of the multiple repetitions of the IG2_MAT_NU region in N. vectensis DG. (A) Multiple sequence alignment of the six IG2_MAT_NU modules of N. vectensis DG compared with this region of human DG. Alignments were prepared in MUSCLE 3.8 and are presented in Boxshade. Asterisks indicate the GS proteolysis site in human DG. At each position, black background includes identical residues; grey background indicates conservative substitutions, and white background indicates non-conserved residues. (B) Phylogenetic analysis of the six IG2_MAT_NU modules of N. vectensis DG (alignment of 153 positions) compared with the same region of human DG. The tree was prepared in PhyML with 200 cycles of boot-strapping. Numbers indicate bootstrap support values, with 1 as maximum. Scale=substitutions/site.