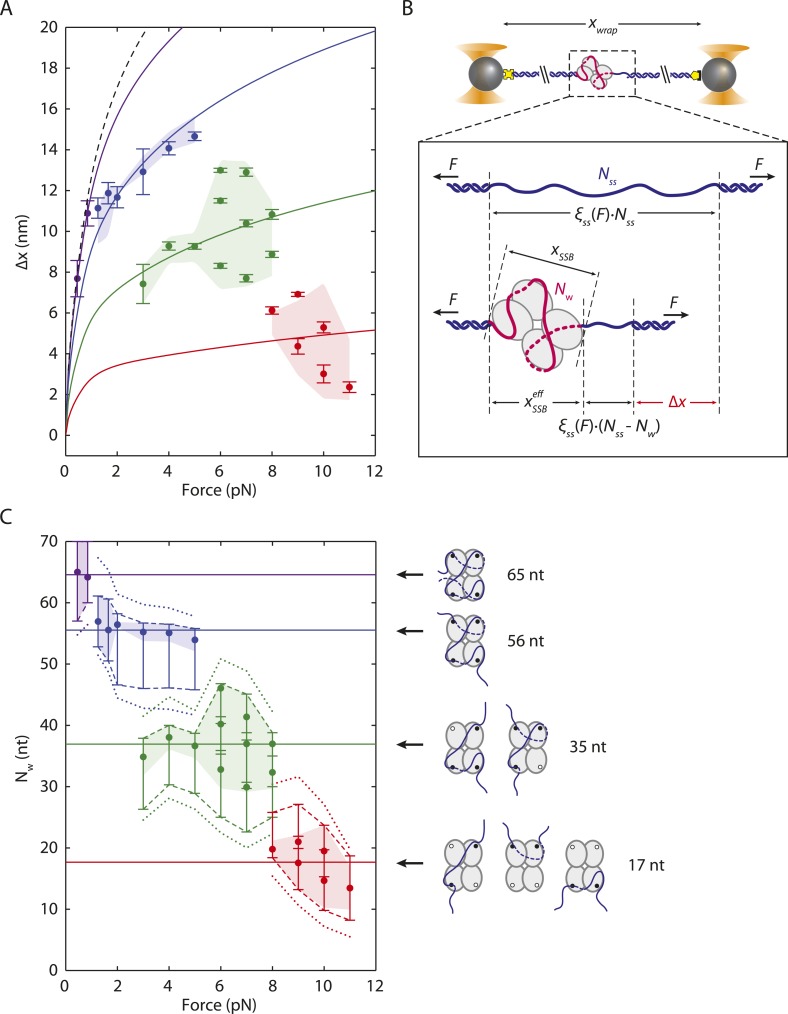
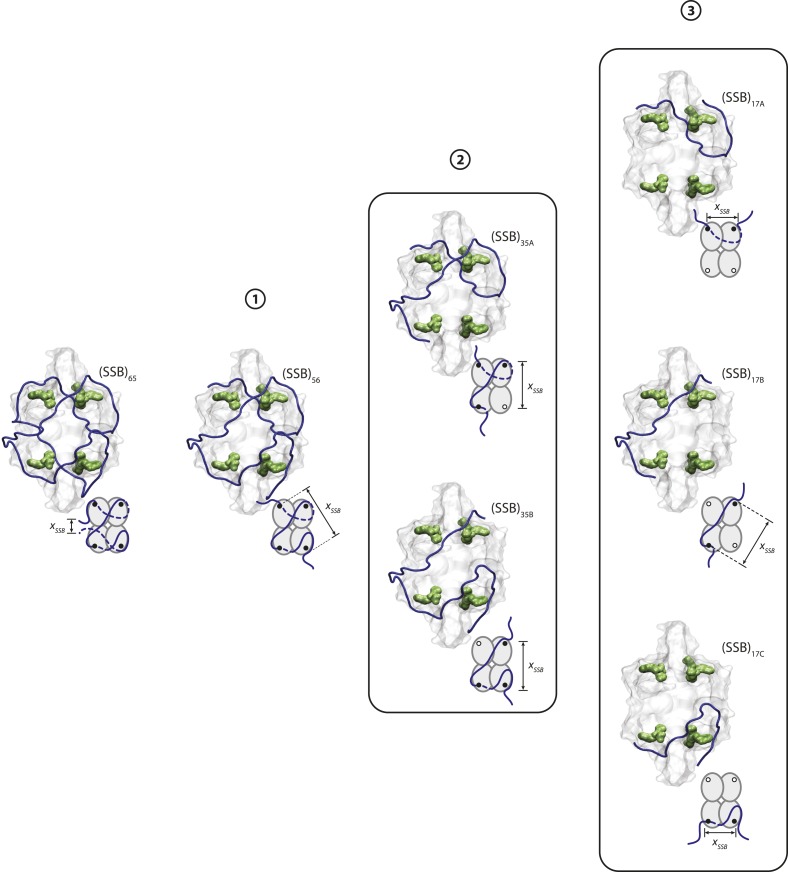
Figure 3. SSB wrapping modes.
(A) Mean change in extension Δx vs tension for each wrapping state, derived from the peaks of the distributions in Figure 2C. Error bars represent S.E.M. and were determined by bootstrapping. The dashed line is the model in Figure 1D. Solid lines represent models of Δx based on Equation 3 for SSB wrapping Nw = 65, 56, 35, and ∼17 nt (purple, blue, green, and red, respectively; ‘Materials and methods’). Data points are clustered into 4 groups corresponding to those states (purple, blue, green, and red circles). (B) Schematic representation of Δx. Top: Bare ssDNA (with Nss = 70 nt) and its extension, xbare, based on a polymer elasticity model Equation 1 (‘Materials and methods’). Bottom: SSB-wrapped ssDNA showing the number of wrapped nucleotides, Nw (<70, red) and the remaining unwrapped nucleotides (Nss − Nw, blue). The extension of wrapped DNA, xwrap is calculated from an elasticity model and the effective physical size of the SSB-ssDNA complex, , Equation 2 (‘Materials and methods’). Δx is the difference between xwrap and xbare, Equation 3. (C) Number of wrapped nucleotides Nw vs tension F. Each data point in (A) is mapped to Nw using the model described in the text (‘Materials and methods’; Figure 3—figure supplement 1). Dotted lines represent the maximum possible range of Nw for each colored group of points based on being <6.5 nm (Figure 3—figure supplement 1, left panel). Dashed lines represent a tighter range of possible Nw for each group of points derived from the SSB-ssDNA structure (Figure 3—figure supplement 1, middle panel). Error bars represent this range for each individual data point. The shaded areas represent the tightest range of possible Nw for each group based on the ‘hotspot’ analysis described in the text (Figure 3—figure supplement 1, right panel). The points are the best estimates of Nw from the model. The shaded areas and solid lines in (C) map directly to those in (A). Cartoon schematics depict possible wrapping modes corresponding to the 4 groups.