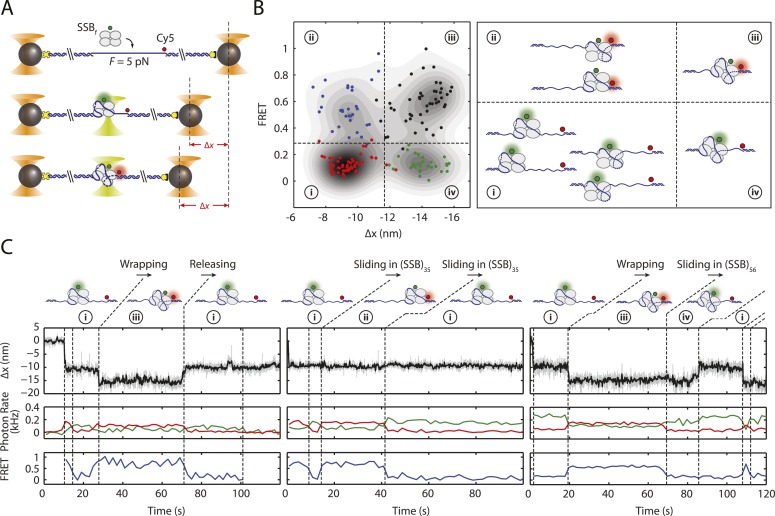
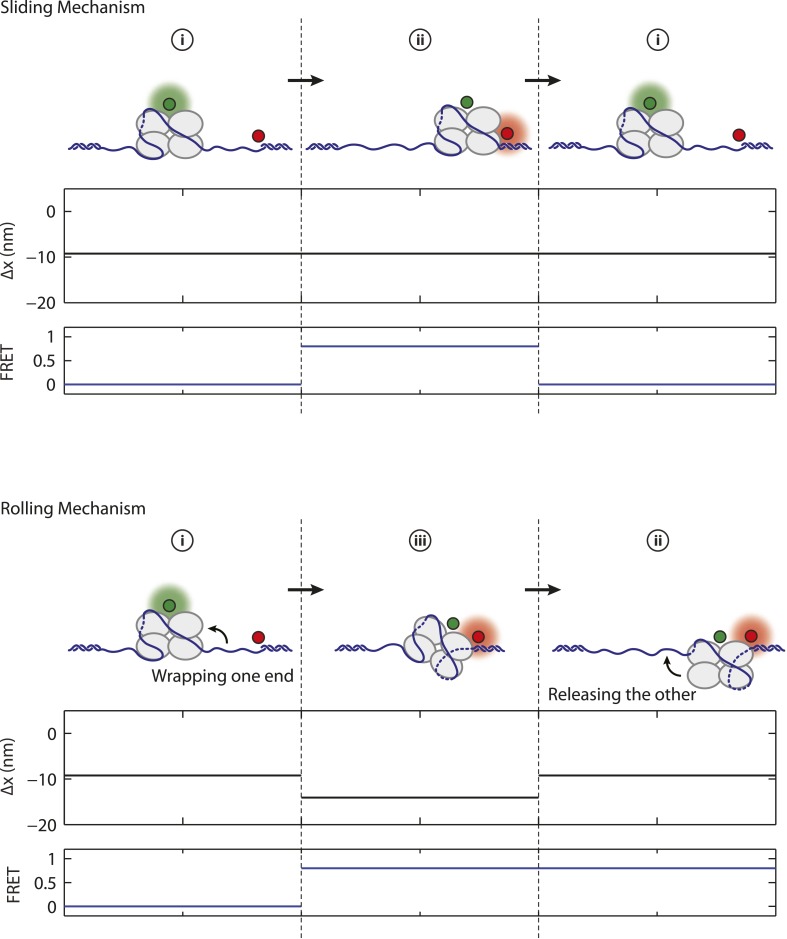
Figure 4. SSB binding modes and diffusion mechanism.
(A) Schematic of fluorescently labeled SSB, SSBf, ssDNA wrapping experiment. A Cy5-labeled DNA construct is tethered between two optical traps under a constant tension of 5 pN. Upon binding of an AlexaFluor555-labeled SSB, both DNA extension change, Δx, and single-molecule FRET are measured simultaneously. (B) Scatter plot of FRET efficiency and Δx. Data (circles) are assigned to 4 states (red (i), blue (ii), black (iii), and green (iv)) based on the value of FRET and Δx. A density map of the combined FRET-extension states overlaid with the scatter plot confirms that the data can be separated into 4 states. Cartoon illustrations of nucleoprotein complexes demonstrate possible SSB wrapping configurations corresponding to the 4 assigned states. (C) Representative traces showing combined fluorescence and DNA extension measurements. Change in extension (top; boxcar averaged to 50 Hz) and fluorescence (middle; boxcar averaged to 0.5 Hz) of donor (SSBf, green) and acceptor (Cy5, red) are measured simultaneously. Together, FRET efficiency (bottom; blue) and extension change (top; black) reveal the SSB wrapping states (i and ii, iii and iv) and their dynamics (ssDNA wrapping/releasing and sliding).