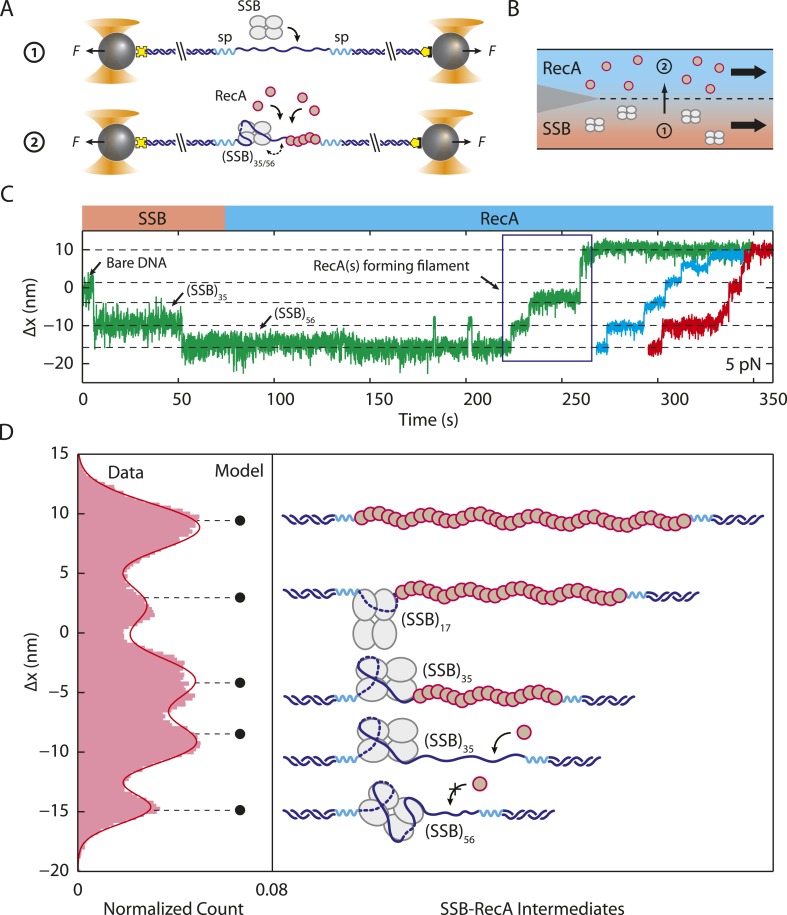
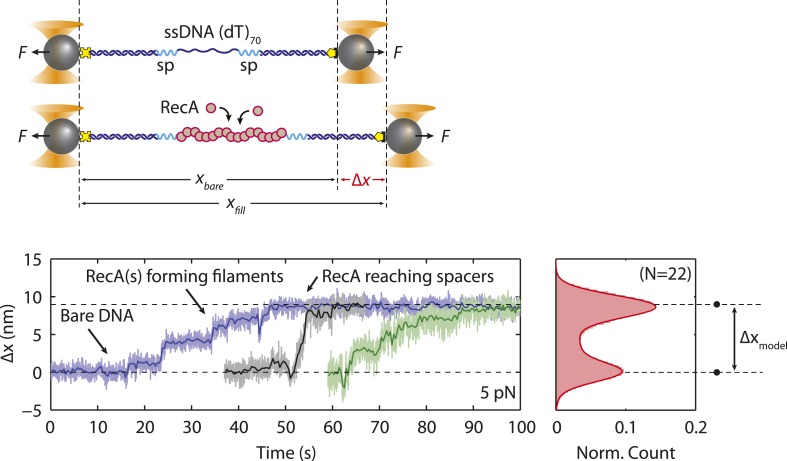
Figure 5. Unwrapping of ssDNA from SSB by RecA filament formation.
(A) Schematic representation of SSB-RecA experiment. A standard DNA construct consisting of a 70-nt single-stranded DNA ((dT)70) fragment was synthesized to contain two internal 18-atom hexa-ethylene-glycol spacers at both ss-dsDNA junctions (cyan; ‘Materials and methods’). The spacers prevent RecA filament formation onto the dsDNA. The construct is tethered in the presence of SSB (Position 1). After the SSB binds, the tethered DNA is moved to the stream containing RecA for observation (Position 2). (B) Experimental flow chamber for SSB-RecA experiment. Two separate streams contain experimental buffer plus 0.5 nM SSB (red, Position 1) and buffer plus 125 nM RecA and 125 μM ATP-γS (blue, Position 2). (C) Representative time traces showing competition between RecA and SSB on ssDNA (green, blue, red). Transient wrapping-unwrapping of SSB slows down the nucleation of RecA. Formation of RecA filament extends ssDNA (blue box), displaces the SSB, and stops after reaching the spacers at the ss-dsDNA junctions. The dotted lines correspond to the model in (D). (D) Extension change distribution of SSB-RecA intermediates at a constant tension of 5 pN (pink) obtained from many RecA filament formation time traces (N = 25). Five states representing SSB-RecA dissociation intermediates are illustrated (schematics) and assigned to peaks of the distribution. Extensions corresponding to these states are predicted using polymer models of elasticity (black dots and dotted lines, ‘Materials and methods’).