Figure 2.
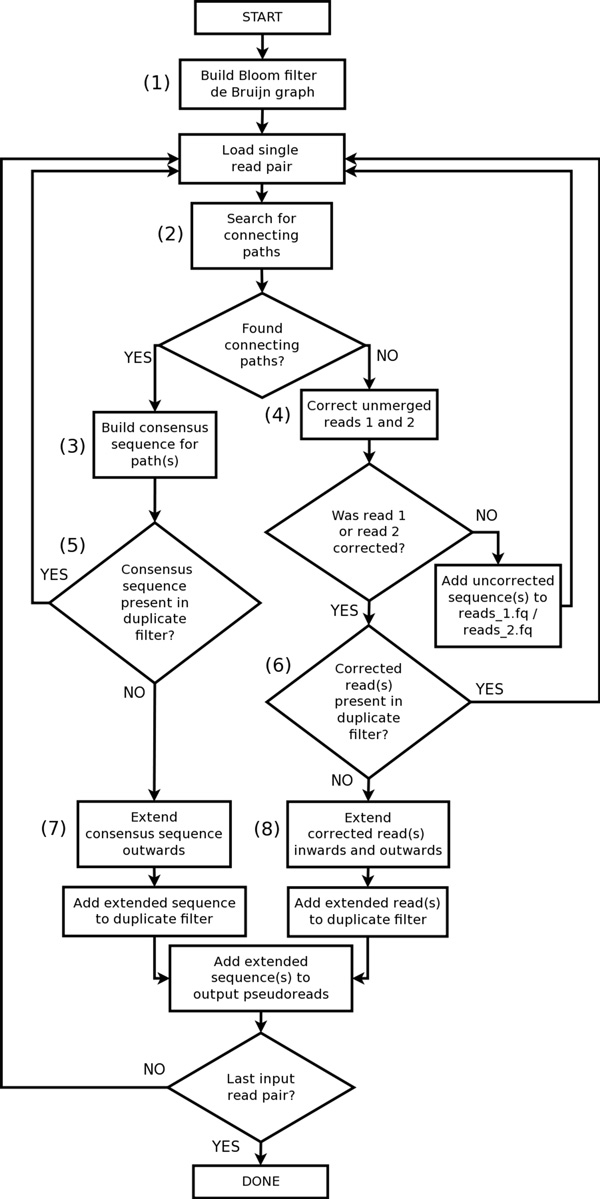
The Konnector2 algorithm. (1): The algorithm builds a Bloom filter representation of the de Bruijn graph by loading all k-mers from the input paired-end sequencing data. (2): For each read pair, a graph search for connecting paths within the de Bruijn graph is performed. (3): If one or more connecting paths are found, a consensus sequence for the paths is built. (4): If no connecting paths are found, error-correction is attempted on reads 1 and 2. (5) and (6): the algorithm queries for the existence of either the consensus connecting sequence or the error-corrected reads in the "duplicate filter". The duplicate filter is an additional Bloom filter, separate from the Bloom filter de Bruijn graph, which tracks the parts of the genome that have already been assembled. (7) and (8): If one or more of the k-mers in the query sequence are not found in the duplicate filter, the sequence is extended outwards in the de Bruijn graph, until either a dead end or a branching point is encountered in the graph. Finally, the extended sequences are written to the output pseudo-reads file.
