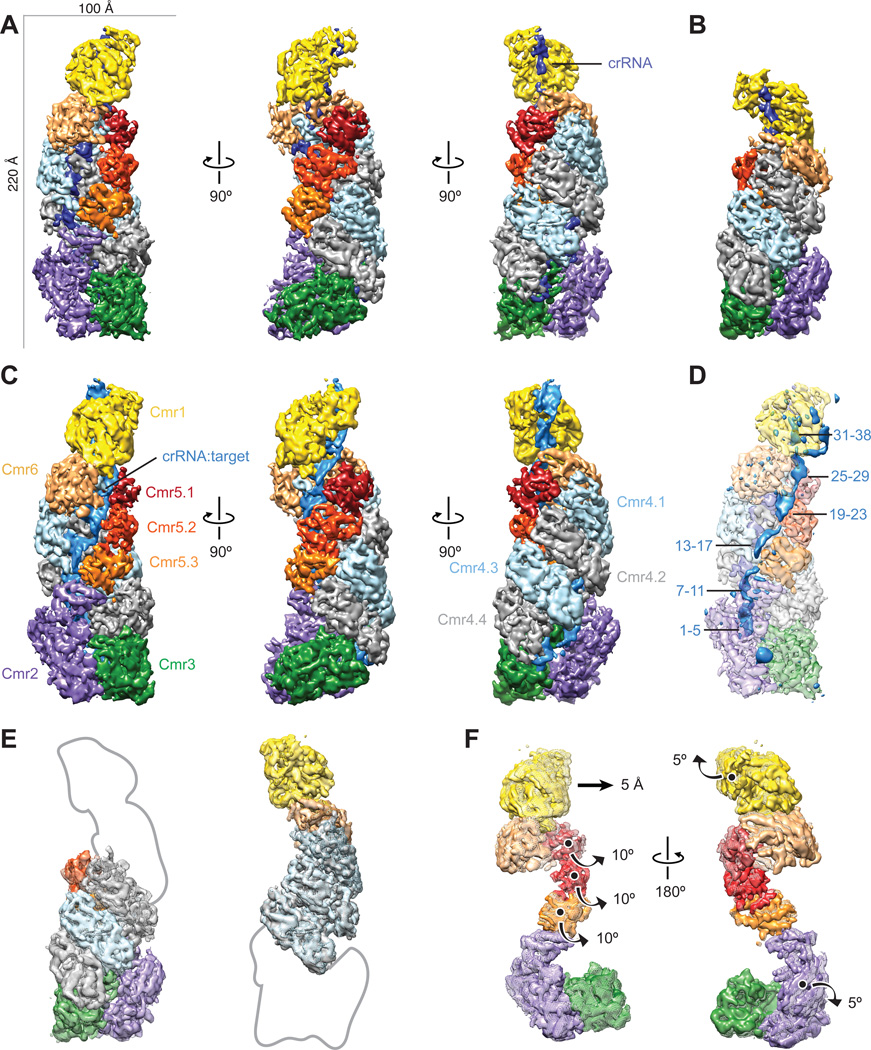
Fig. 1. Architecture of the native crRNA-bound (apo) and ssRNA target-bound Cmr.
(A–C) Cryo-EM reconstructions of intact apo-Cmr (crRNA-bound) (A), a smaller apo-Cmr (B), and ssRNA target-bound Cmr (C), at 4.1-, 4.5- and 4.4-Å resolution (using the 0.143 gold standard Fourier Shell Correlation criterion), respectively. Subunits are segmented and colored as indicated. (D) Difference map between intact apo-Cmr and target-bound Cmr at 10-σ (solid, blue) superimposed on the apo-Cmr structure (transparent). (E) Aligning the smaller (surface) and intact (mesh) apo-Cmr complexes based on Cmr2–Cmr3 (left) or Cmr1–Cmr6 (right), shows that the helical geometry is preserved. (F) Aligning apo-Cmr (surface) and target-bound Cmr complexes (transparent mesh) based on the Cmr4 backbone (removed for clarity), reveals this surveillance complex undergoes concerted conformational changes upon ssRNA substrate recognition. Details of these movements can be seen in Movie S1, S2.