Fig. 3.
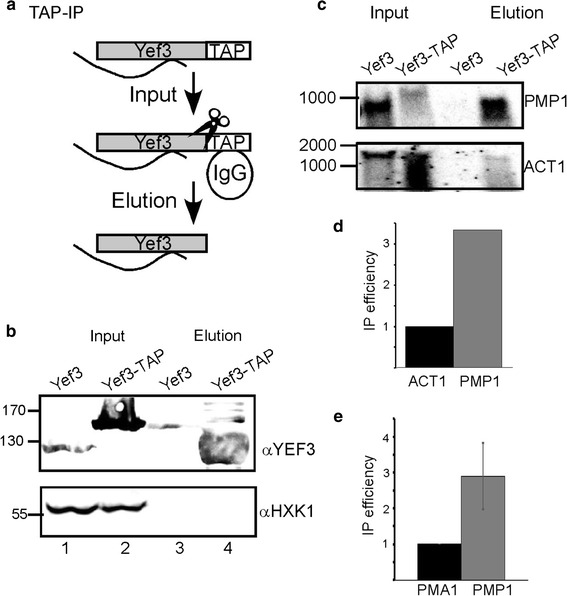
Validation of RaPID results by TAP purification. a TAP protocol. Cells expressing TAP-tagged Yef3 are lysed and loaded on IgG-Sepharose beads (input). The beads are washed extensively from non-specific binders and Yef3 is specifically cleaved off the beads by a TEV protease (depicted as scissors). Eluted material is analyzed for proteins (panel b) or mRNAs (panels c–e). b Western analysis with the indicated antibodies for aliquots from the Input and Elution. Note that the TAP tagged Yef3 is larger by ~25 kDa from the normal Yef3. Thus, the signal in lane 3 is due to spill over from lane 2 and not non-specific association of untagged Yef3. c Northern analysis for Input and Elution RNA samples isolated from a strain expressing untagged Yef3 or TAP-tagged Yef3. Membranes were hybridized with the probes indicated to the right. RNA size markers are indicated to the left. d Averages of two independent northern analyses. Results are presented as the ratio of Elution to Input samples, normalized to ACT1 efficiency. e RT-qPCR quantitation of the amounts of PMA1 and PMP1, isolated from the tagged (Yef3-TAP) strains. Results are presented as the ratio of Elution to Input, normalized to the PMA1 results. Results are from at least 3 independent biological repeats, from cell growth through IP to RT-qPCR analysis. Error bars are SEM of three independent biological repeats
