Background: OsGAP1, a plant-specific C2-domain protein, binds to phospholipids and an unconventional G-protein (OsYchF1).
Results: We solved the OsGAP1 structure and performed site-directed mutagenesis to identify two functional surfaces for binding to phospholipids versus OsYchF1.
Conclusion: Interactions with phospholipids and OsYchF1 play important roles in the functions of OsGAP1.
Significance: Our data advance the understanding of the structure-function relationship of C2-domain proteins.
Keywords: crystal structure, G protein, phospholipid, plant defense, plasma membrane
Abstract
The C2 domain is one of the most diverse phospholipid-binding domains mediating cellular signaling. One group of C2-domain proteins are plant-specific and are characterized by their small sizes and simple structures. We have previously reported that a member of this group, OsGAP1, is able to alleviate salt stress and stimulate defense responses, and bind to both phospholipids and an unconventional G-protein, OsYchF1. Here we solved the crystal structure of OsGAP1 to a resolution of 1.63 Å. Using site-directed mutagenesis, we successfully differentiated between the clusters of surface residues that are required for binding to phospholipids versus OsYchF1, which, in turn, is critical for its role in stimulating defense responses. On the other hand, the ability to alleviate salt stress by OsGAP1 is dependent only on its ability to bind OsYchF1 and is independent of its phospholipid-binding activity.
Introduction
Eukaryotic membrane systems play crucial roles in cellular processes. To facilitate the targeting of regulatory proteins to specific membranes and cellular compartments, various lipid-binding domains have evolved (1–4). There are at least 11 lipid-binding domains, among which the eukaryote-specific C2 domain is the second most abundant (3). It is characterized by a β-sandwich structure connected by surface loops comprising eight primary β-strands and one α-helix that may exist in a conserved position among the same family of C2 domain (4, 5). Most C2-domain proteins are soluble and can bind reversibly to membranes, whereas some others are trans-membrane proteins (3). The low primary sequence homology among extant C2 domains points to the possibility that they may have diversified before the appearance of eukaryotes (4).
In animals, C2 domains are found in diverse regulatory proteins that play central roles in cellular physiology, such as protein kinase Cs (PKCs), phospholipases, lipoxygenases, synaptotagmins, GTPase-activating proteins (GAPs),3 etc. (1–4).
In plants, there are at least four distinct groups of C2-domain proteins (6), including a plant-specific group of small proteins with a single C2 domain. Functional studies of small C2-domain proteins in plants have indicated their roles in pollen fertility (7) as well as abiotic stress and plant defense responses (8, 9). However, the importance of the structure-functional role of small C2 proteins was not explored until a recent report on the Arabidopsis thaliana AtCAR4 protein (10), which is identical to the Arabidopsis homologue AtGAP1 that we have reported previously (11). AtCAR4 directs the interaction of abscisic acid receptors with the plasma membrane and hence is involved in the sensitivity of this signal pathway.
Our target C2-domain protein, the rice OsGAP1, is a unique member of this group of plant-specific small C2 proteins. OsGAP1 is a soluble protein that can loosely associate with plasma membrane, presumably via reversible binding to phospholipids and therefore shuttle between the cytosol and the plasma membrane (12). In addition, it can also interact with an unconventional G-protein, OsYchF1, and stimulate its NTPase activities (12), thus representing the first example of its kind of small C2 proteins. Furthermore, it plays a positive role in both plant defense and abiotic stress responses, presumably through its interactions with phospholipids and OsYchF1 (11–13). However, the detailed mechanisms of these interactions in relation to its structure and functions remain a mystery.
Here, we determined the structure of OsGAP1 by x-ray crystallography. Based on the structure and sequence analyses, we have generated mutants of OsGAP1 to map the regions responsible for binding phospholipids and OsYchF1. Using the site-directed mutagenesis approach and our established gain-of-function transgenic A. thaliana testing model (11–13), we were able to demonstrate the significance of binding to phospholipids and to OsYchF1 for the functional roles of OsGAP1.
Experimental Procedures
Protein Expression for X-ray Crystallography
The OsGAP1 cDNA was amplified from a rice (Oryza sativa) cDNA library by polymerase chain reaction (PCR) and inserted into the EcoRI/BamHI restriction sites of the pBluescript II SK(+) vector. Subsequently, the AgeI-HindIII-digested OsGAP1 cDNA fragment was subcloned into an expression vector, pRSETA-HisSUMO (14), and transformed into the Escherichia coli strain BL21 (DE3). Protein expression was induced by adding 1.0 mm isopropyl β-d-thiogalactopyranoside. The cell pellets were used immediately or stored at −80 °C. Cell pellets were resuspended in 35 ml of buffer A (20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 50 mm imidazole). After sonication, the cell-free extract was applied onto a buffer A-equilibrated Ni2+-chelating resin column. After removing the unbound E. coli proteins, the fusion proteins were eluted with buffer B (20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 300 mm imidazole). The HisSUMO tag was then removed by HisSUMO protease, followed by gel filtration with Sephadex 75 in a running buffer (20 mm Tris-HCl, pH 7.3, 100 mm NaCl).
Crystallization Data Collection and Structure Determination
The x-ray-quality OsGAP1 crystals were grown at 16 °C by the sitting drop vapor diffusion method using a medium comprising 0.1 m potassium sodium tartrate tetrahydrate and 20% (w/v) PEG 3350. Crystals were equilibrated in a cryoprotectant including the mother reservoir solution supplemented with 15% glycerol. The diffraction data were collected on an in-house diffractometer (Rigaku FR-E+), indexed, and integrated using the iMosfilm program (15). The phase problem was solved by molecular replacement using the C2B domain of rabphilin-3A (Protein Data Bank entry 2CM6) as a search model (16), and data were processed using the CCP4 program suite. The model was built and refined with Coot (17) and Phenix (18) and visualized by PyMOL (PyMOL Molecular Graphics System, Schrödinger, LLC, New York). The data collection statistics and refinement statistics are listed in Table 1.
TABLE 1.
Summary of statistics for diffraction data collection and structure refinement
Values in parentheses are for the highest resolution shell.
| OsGAP1 | |
|---|---|
| Data collection | |
| Space group | C 2 2 21 |
| Cell dimensions | |
| a, b, c (Å) | 37.19, 166.75, 60.73 |
| α, β, γ (degrees) | 90, 90, 90 |
| Wavelength (Å) | 1.5418 |
| Resolution (Å) | 30.37–1.63 (1.72–1.63) |
| Rmerge | 0.106 (0.307) |
| I/Σi | 12.3 (5.0) |
| Completeness (%) | 98.9 (97.4) |
| Redundancy | 6.9 (6.4) |
| Refinement | |
| Resolution (Å) | 30.37–1.63 (1.67–1.63) |
| No. of reflections | 23480 (1562) |
| Rwork/Rfree | 0.1715/0.1977 (0.2534/0.2904) |
| No. of atoms | |
| Protein | 1265 |
| Solvent | 327 |
| B-Factors | |
| Protein | 18.26 |
| Water | 31.08 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (degrees) | 1.306 |
| Ramachandran analysis | |
| Preferred (%) | 98.10 |
| Allowed (%) | 1.90 |
Construction of OsGAP1 Mutants by Site-directed Mutagenesis
Overlapping PCR (19) was performed to generate constructs with targeted mutations in the surface cluster regions that are predicted to interact with either proteins or phospholipids. The primers used were listed in Table 2. Two rounds of PCR were performed; fragments with the N-terminal half and the C-terminal half of OsGAP1 with overlapping mutated regions were generated separately in the first round of PCR. The two fragments were then mixed and further amplified to generate full-length mutated constructs in the second round of PCR. The PCR products were then inserted into the EcoRI and SalI sites of the pMAL-c2 expression vector to generate fusion constructs with the maltose-binding protein (MBP) (20). DNA sequencing was performed to confirm the success in mutagenesis and cloning.
TABLE 2.
Primers used in site-directed mutagenesis of OsGAP1
| Description | Sequences (5′ to 3′) |
|---|---|
| Universal primers used as outer boundary primers to amplify each of the modified OsGAP1 except Cluster 1 mutanta | |
| Forward primer | CCGAATTCATGTTGGGGCATCTGGTTG |
| Reverse primer | AACTCGAGAATAGGCAGAGTACAGAGTTC |
| Outer primer to introduce part 1 of Cluster 1 mutationsb | |
| Forward primer | CCGAATTCATGTTGGGGCATGCGGTTGGGGCGGTGAAG |
| Overlapping primer pair to introduce part 2 of Cluster 1 mutations | |
| Forward primer | GATGAACTCGCCCTCGCGATCGAAGAT |
| Reverse primer | ATCTTCGATCGCGAGGGCGAGTTCATC |
| Overlapping primer pair to introduce Cluster 2 mutations | |
| Forward primer | GTCCGCGCCCTCCGCTCCAGCGCCCCCTAC |
| Reverse primer | GTAGGGGGCGCTGGAGCGGAGGGCGCGGAC |
| Overlapping primer pair to introduce Cluster 3 mutations | |
| Forward primer | ATGGGCGCACAGGCATTGGCAACAGCAGTCATA |
| Reverse primer | TATGACTGCTGTTGCCAATGCCTGTGCGCCCAT |
| Overlapping primer pair to introduce Cluster 4 mutations | |
| Forward primer | AATGCACAGGCCTGCCTAGCTGCAGCGAGC |
| Reverse primer | GCTCGCTGCAGCTAGGCAGGCCTGTGCATT |
| Overlapping primer pair to introduce Cluster 5 mutations | |
| Forward primer | GTTCTCGCACTAGCGGATGTGGCATGCGGGGCAATT |
| Reverse primer | AATTGCCCCGCATGCCACATCCGCTAGTGCGAGAAC |
a EcoRI and XhoI recognition sites were added to the 5′ ends of the primer pair to facilitate the subsequent cloning steps.
b No overlapping primers were needed for the mutations in Cluster-1 (part 1) because the mutation region was located within the outer forward primer priming site. Mutation was directly achieved by designing mutations at the outer forward primer and then pairing it with the overlapping reverse primer of the mutations in Cluster-1 (part 2) to produce the mutated N-terminal OsGAP1 fragment.
Expression and Purification of MBP Fusion Proteins of the Wild Type and Mutant OsGAP1s and the GST Fusion Protein of OsYchF1
After the pMAL-C2 constructs were transformed into the E. coli BL21 (DE3) strain, protein expression was induced by adding 0.5 mm isopropyl β-d-thiogalactopyranoside in LB medium with 100 mg/liter ampicillin and grown at 20 °C overnight. SpinCleanTM MBP Excellose® spin kit (Mbiotech 23020) was then used to purify the expressed proteins following the procedures described in the user manual. The glutathione S-transferase (GST) fusion protein of OsYchF1 was obtained by expressing the construct in the pGEX-4T-1 expression vector as described in a previous report (12) and purified by the MagneGSTTM protein purification system (Promega V8603), following the procedures described in the user manual.
Analysis of Protein-Phospholipid Interaction by Dot Blot Assays
For preliminary screening of phospholipid affinities of OsGAP1 and its mutants, phospholipid dot blot assays were accomplished by dotting 1 μl of phospholipid mixture from soybean (Supelco P3817-1VL, containing l-α-lysophosphatidylcholine (0.3 mg/ml), l-α-phosphatidylcholine (1.5 mg/ml), l-α-phosphatidylethanolamine (1.2 mg/ml), and l-α-phosphatidylinositol sodium salt (0.9 mg/ml)) on a supported nitrocellulose membrane (Bio-Rad 162-0095). The membrane was then blocked with 3% skim milk and incubated with 0.2 mg/ml (in excess) MBP fusion proteins of the wild type or mutant OsGAP1 in the blocking solution and washed as described previously (12). Tris-buffered saline supplemented with 0.1% (v/v) Tween 20 was used for the subsequent incubation and washing steps. The mouse monoclonal anti-MBP antibody (Sigma M6295) and the sheep anti-mouse HRP-linked secondary antibody (GE Healthcare, NA931) were used for detection. Biological repeats were performed to confirm the results.
The phospholipid-binding specificities of MBP-fused wild type OsGAP1 and Clusters 1, 2, 3, and 4 mutants were further examined using membrane lipid strips (Echelon, P-6002). Each strip was predotted with 15 different biologically important membrane lipids, including triglyceride, phosphatidylinositol (PtdIns), PtdIns 4-phosphate, PtdIns 4,5-bisphosphate, PtdIns 3,4,5-trisphosphate, phosphatidylserine, phosphatidylethanolamine, phosphatidic acid, diacylglycerol, cholesterol,phosphatidylcholine, sphingomyelin, phosphatidylglycerol, 3-sulfogalactosylceramide, and cardiolipin. The protocol suggested in the manufacturer's manual was followed, except that all blocking and washing buffers were supplemented with 200 μm CaCl2. Mouse monoclonal anti-MBP antibody (Sigma, M6295) and the sheep anti-mouse HRP-linked secondary antibody (GE Healthcare, NA931) were used for detection. Biological repeats were performed to confirm the results.
Circular Dichroism Measurement of Native and Mutated OsGAP1 Protein
Circular dichroism measurements were performed with a JASCO J-815 spectrometer using a 1-mm rectangular quartz cuvette. The spectra of all samples with a protein concentration of 0.2 mg/ml were collected in the dark at 25 °C.
In Vitro Pull-down Experiment
E. coli cell lysates containing the MBP fusion proteins of the wild type or mutant OsGAP1 were added to GST-OsYchF1-bound MagneGSTTM glutathione particles from the MagneGSTTM protein purification system (Promega, V8603). The mixture was washed and eluted according to the user manual, followed by Western blot. The presence of MBP-OsGAP1 fusion proteins was detected by the mouse monoclonal anti-MBP antibody (Sigma, M6295), followed by the goat anti-mouse AP-linked secondary antibody (Sigma, A0162). Color detection was done after adding the SIGMA FASTTM 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium substrate (Sigma, B5655). Biological repeats were performed to confirm the result.
Growth Conditions of A. thaliana
Floragard universal potting soil from Floragard Vertriebs GmbH (Gerhard-Stalling, Germany) was used for the cultivation of A. thaliana in a growth chamber (22 °C, 70–80% relative humidity, light intensity of 80–120 μmol m−2 s−1 on a 16-h light/8-h dark cycle).
Preparation of Transgenic A. thaliana
To constitutively express the wild type and mutant OsGAP1 proteins in A. thaliana, the recombination constructs were subcloned into a binary vector (V7) (21) and expressed under the control of the cauliflower mosaic virus 35S promoter before being transformed into the wild type Arabidopsis (Col-0) via a vacuum infiltration method (22) using the Agrobacterium tumefaciens strain GV3101 (pMP90) (23). Screening of the T1 seeds was performed on MS agar plates supplemented with 50 mg/liter kanamycin. A phenotypic ratio of kanamycin resistance of 3:1 shown in the T2 generation indicated a single-locus insertion event. Homozygous lines screened from the T3 generation were used in functional experiments. Real-time RT-PCR was performed to confirm the expression of the transgenes.
Biotic and Abiotic Treatments of Transgenic A. thaliana
Transgenic and wild type (Col-0) Arabidopsis lines were subjected to biotic (Pseudomonas syringae pv. tomato (Pst) DC3000 inoculation) and abiotic (150 mm NaCl) stresses. Syringe infiltration of Pst DC3000 was performed on the abaxial side of leaves on 6-week-old plants (24). Tissue samples were harvested on days 0 and 3 for colony-forming unit estimation and RNA extraction. Real-time RT-PCR was used to determine the expression levels of defense marker genes (PR1 and PR2) (12, 13). For abiotic stress experiments, 10-day-old seedlings were transferred onto MS agar plates supplemented with 150 mm NaCl. Chlorophyll contents were measured as described (25, 26). Real-time RT-PCR was used to examine the expression levels of salt stress-responsive genes (RD22 and RD29a) from tissue samples harvested 1 day after treatment (11).
GTPase Activity Assay
The effects on the GTPase activity of OsYchF1 by the wild type and mutant OsGAP1 proteins were monitored via the release of inorganic phosphate (Pi) during GTP hydrolysis, using the EnzChekTM phosphate assay kit (E6646; Molecular Probes, Inc., Carlsbad, CA) (27). To measure the GTPase-activating activities, 10 ng of MBP fusion proteins of wild type or mutant OsGAP1 was mixed with 25 ng of GST-OsYchF1. The GTPase activities were measured in the presence of 200 μm GTP. The wild type MBP-OsGAP1 alone was also tested to confirm that there was no intrinsic GTPase activity in OsGAP1. The MBP protein alone was employed as the negative control.
Statistical Analyses
Statistical analyses were performed using the IBM Statistical Package for Social Sciences (SPSS) (version 22.0). The mean differences were analyzed using one-way analysis of variance followed by the Games-Howell or Tukey's post hoc tests.
Results
Crystal Structure of OsGAP1
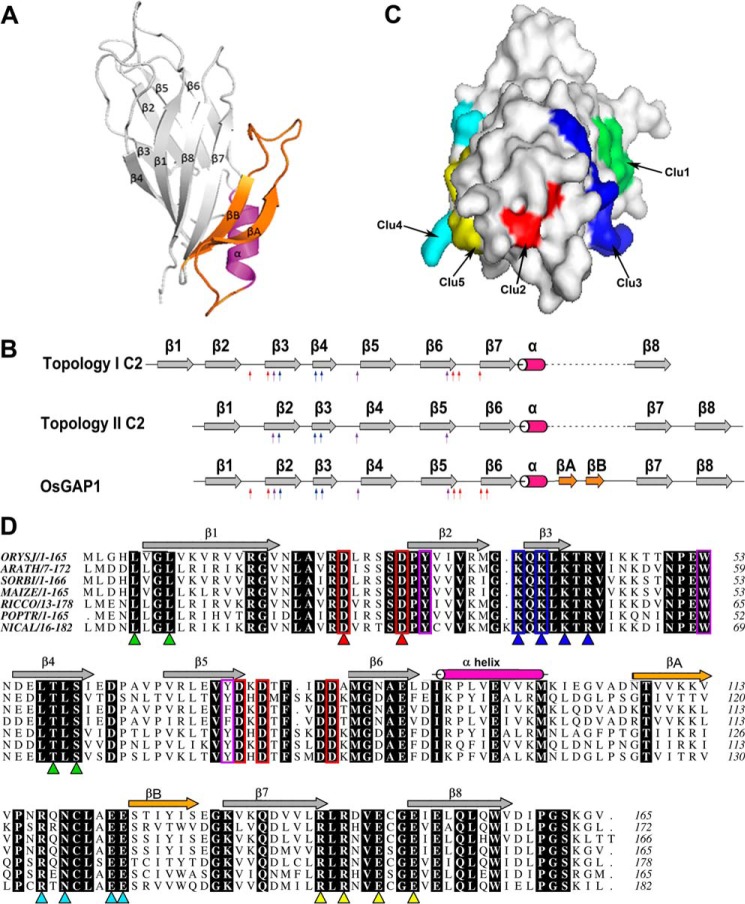
The crystal structure of OsGAP1 was obtained to a resolution of 1.63 Å (Fig. 1A; Protein Data Bank entry 4RJ9). OsGAP1 has two four-stranded anti-parallel β-sheets that form a sandwich structure typical of a C2 domain (4, 5, 28). Among the seven C2-domain families reported (4), the sequence of OsGAP1 is closest to the PKC-C2 family. The orders of the β-strands are in 4-1-8-7 and 3-2-5-6 topologies that resemble the type II topology of a C2-domain fold (28) (Fig. 1B). On the other hand, there are two unique structural features of OsGAP1 that distinguish it from other families of C2-domain proteins: (i) there are two additional β-strands (βA and βB) inserted between β6 and β7, and (ii) OsGAP1 contains only one single C2 domain (Fig. 1).
FIGURE 1.
Structural analysis of OsGAP1. A, a ribbon diagram showing the structure of OsGAP1 as resolved by x-ray crystallography to a resolution of 1.63 Å. B, a schematic diagram showing that OsGAP1 exhibits overall a type II C2-domain topology but possesses signature residues conserved among type-I C2 domains. Red arrows indicate conserved aspartate residues in the putative lipid-binding loop typical of type I topology. Blue and purple arrows, positively charged residues in the putative cationic β-groove and conserved aromatic residues, respectively (conserved in both topologies). Two unique β-strands newly discovered only in OsGAP1 (βA and βB) are indicated in orange. C, a surface diagram showing the locations of the five clusters of mutated residues. Clusters 1 and 3 (Clu1 and Clu3) are closer in one face, whereas Clusters 4, 5, 2, and 3 (Clu4, Clu5, Clu2, and Clu3) line up at another angle. D, sequence alignment of plant homologues of OsGAP1. Identical residues among homologues are highlighted in black. Five clusters of amino acids designated for mutagenesis are indicated by triangles (green, Cluster 1; red, Cluster 2; blue, Cluster 3; cyan, Cluster 4; yellow, Cluster 5). The main criteria for choosing these residues were that they must be surface residues and must be conserved among other plant GAP1 homologues. Conserved residues common to the PKC-C2 domain are boxed (red, aspartate residues making up a calcium-binding pocket; blue, lysine patch; purple, aromatic residues). Five clusters of residues were chosen as the targeted mutation sites. ORYSJ, homologue from O. sativa subsp. Japonica (GenBankTM accession number NP_001046709); it is the same gene encoding OsGAP1. ARATH, homologue from A. thaliana (GenBankTM accession number NP_188425); it is the same gene encoding AtCAR4. SORBI, homologue from Sorghum bicolor (GenBankTM accession no. XP_002455977). MAIZE, homologue from Zea mays (GenBankTM accession number NP_001132168). RICCO, homologue from Ricinus communis (GenBankTM accession number XP_002530486). POPTR, homologue from Populus trichocarpa (GenBankTM accession number XP_006383322). NICAL, homologue from Nicotiana alata (GenBankTM accession number ACD40010).
While this manuscript was in preparation, Rodriguez et al. (10) published the structure of a homologue of OsGAP1, AtCAR4, from Arabidopsis (Protein Data Bank entry 4V29). The structure of OsGAP1 is superimposable with that of AtCAR4, with a Cα root mean square deviation of 0.84 Å. Both proteins contain the conserved aromatic and lysine residues forming a cationic β-groove responsible for phospholipid-binding specificity (29, 30) (Fig. 1C). They also possess all of the conserved aspartate residues found uniquely among the PKC-C2 members with type I topology (30), assumed to be important for Ca2+ binding and phospholipid interaction (31, 32).
The alignment of OsGAP1 plant homologues identified five clusters of conserved residues on the surface of the three-dimensional structure of OsGAP1 (Fig. 1, C and D). Cluster 1 (Leu-5, Leu-8, Thr-58, and Ser-60) is located on strands β1 and β4. Cluster 2 (Asp-23 and Asp-28) contains two aspartate residues located on the loop connecting strands β1 and β2 (including the signature aspartate residues of the topology I PKC-C2 members). Four basic residues on the β3 strand constitute Cluster 3 (Lys-37, Lys-39, Lys-41, and Arg-43) which includes the conserved lysine residues forming the β-groove of the PKC-C2 members. Cluster 4 (Arg-117, Asn-119, Glu-123, and Glu-124) is located on the loop between the novel strands βA and βB, and Cluster 5 (Arg-141, Arg-143, Glu-146, and Glu-149) is between strands β7 and β8.
Different Amino Acid Residue Clusters of OsGAP1 Are Responsible for Binding Phospholipids and OsYchF1
We previously showed that OsGAP1 can bind to both phospholipids and the unconventional G-protein, OsYchF1 (12, 13). To determine the relative contributions of various clusters of amino acid residues to these two binding abilities, we constructed five mutants. By comparing with other plant homologues of OsGAP1, conserved amino acid residues on the three-dimensional protein structure were identified. Five clusters of residues were chosen as the targeted mutation sites (Fig. 1). The main criteria for choosing these residues were that they must be surface residues and be conserved among other plant OsGAP1 homologues. Other criteria, such as polarity or hydrophobicity, were also taken into consideration. Because most bindings require the amino acid side chains for interactions, all of the residues in the clusters indicated (Fig. 1) were mutated to alanine to remove the effects of the side chains on the interactions.
Two nonpolar aliphatic residues on β-strand 1 and two uncharged polar residues on β-strand 4 constitute Cluster 1. Two aspartate residues in the calcium binding pockets were mutated in Cluster 2. Not all of the residues in the calcium binding pocket were mutated because the resulting total change in charge may be too large and may affect the protein in ways other than the side chain effect. Positively charged lysine and arginine on β-strand 3, which form the cationic β-groove, were mutated in Cluster 3. Last, four charged residues on the loop between β-strand 7 and 7a and on the loop between β-strand 7b and 8 were mutated as Cluster 4 and Cluster 5, respectively (Table 3). To minimize the effects of electrostatic attraction dominating the mutation analysis, the overall changes in charges for the mutated surfaces were kept mostly neutral, except for Clusters 2 and 3 (Table 3), due to their unique roles in the C2 domain. Cluster 2 consists of two aspartate residues that are conserved among the PKC-C2 domain. Mutation of Cluster 2 causes a decrease of two negative charges. Cluster 3 consists of residues forming the cationic β-groove presumably responsible for phospholipid binding specificity. The mutated surface has a decrease of 4 positive charges.
TABLE 3.
Summary of the mutation sites in different surface clusters
| Mutation points | Overall charge changes | Phospholipid binding | OsYchF1 binding/activation | ||||
|---|---|---|---|---|---|---|---|
| Cluster 1 | L5A | L8A | T58A | S60A | 0 | Retained | Lost |
| Cluster 2 | D23A | D28A | +2 | Lost | Retained | ||
| Cluster 3 | K37A | K39A | K41A | R43A | −4 | Partly lost | Lost |
| Cluster 4 | R117A | N119A | E123A | E124A | 0 | Lost | Retained |
| Cluster 5 | R141A | R143A | E146A | E149A | 0 | Lost | Retained |
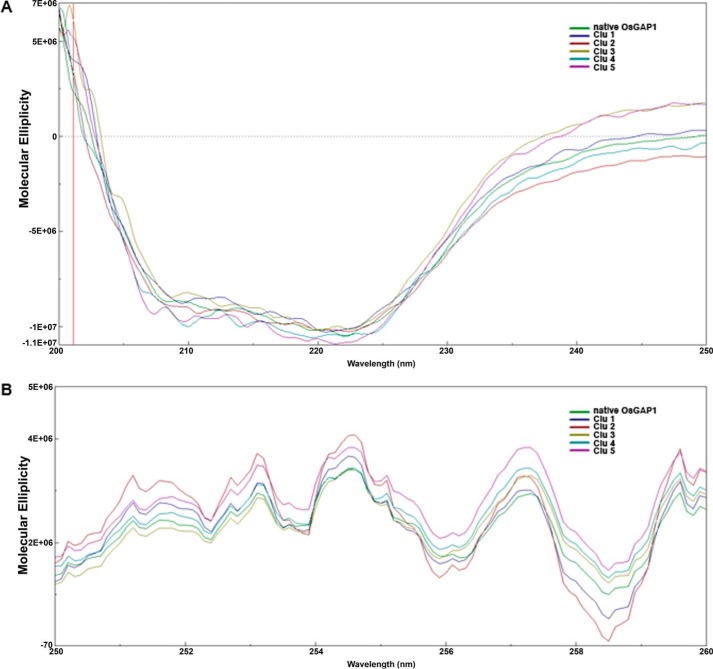
MBP fusions of the wild type and mutant OsGAP1s were successfully expressed and purified. Effects of amino acid changes in the five clusters on protein folding were monitored by circular dichroism analysis. In the far UV region, the circular dichroism spectra of OsGAP1 mutants were similar to that of the wild type protein with a negative value between 203 and 245 nm (Fig. 2), indicating that the secondary structures (i.e. β strands) of the mutants were properly folded just like the wild type protein was (33). In addition, the near UV spectra of the mutants can be well superimposed on that of the wild type protein (Fig. 2). This implied that the microenvironments between the side chains of aromatic residues on all of the proteins were similar (33). Therefore, all mutants were folded properly to maintain an intact protein structure. No significant distortion of structure was seen in the mutants as compared with the wild type OsGAP1. Any loss in function could then be attributed to the removal of the side chains in the mutated residues but not the distortion of the wild type structure.
FIGURE 2.
Circular dichroism spectra of native OsGAP1 and its mutants (Clu1 to -5) in the far UV region (A) and the near UV region (B). Spectrum with buffer only was also measured for normalization. Native MBP-OsGAP1 was included for comparison. Clu1, MBP-OsGAP1 with Cluster 1 mutated; Clu2, MBP-OsGAP1 with Cluster 2 mutated, etc. Molecular ellipticities were calculated via fine tuning with the exact protein concentration in the cuvette.
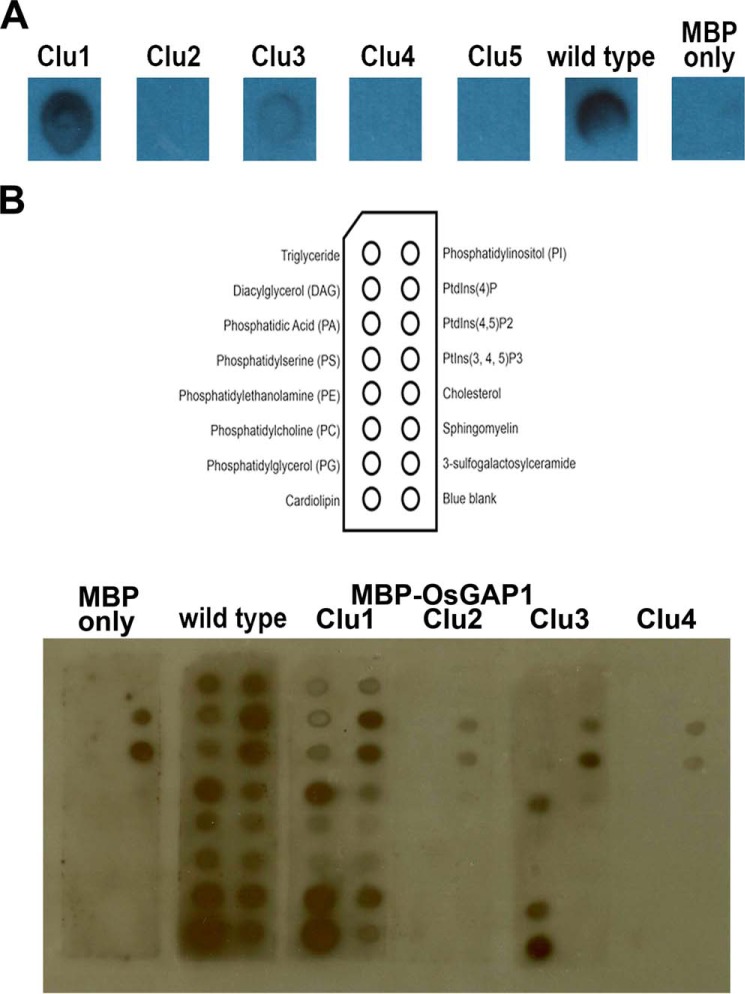
The binding to phospholipids was first investigated qualitatively by dot blot assays using a commercial soybean phospholipid mixture (Fig. 3A). Mutations in Cluster 1 did not affect phospholipid binding, whereas mutations in Cluster 3 weakened this binding. On the other hand, mutations in Clusters 2, 4, and 5 abolished phospholipid binding altogether (Fig. 3A).
FIGURE 3.
Site-directed mutagenesis of OsGAP1 to dissect binding to phospholipids. A, phospholipid dot blots of the wild type and mutant OsGAP1s. One μl each of the phospholipid mixture from soybean (Supelco) was dotted on supported nitrocellulose membranes (Bio-Rad). The membranes were then incubated with purified MBP-OsGAP1 proteins. The interaction was detected by anti-MBP antibodies (Sigma M6295). Mutating Cluster 1 did not affect the phospholipid binding activity of OsGAP1, but mutations in Clusters 2–5 abolished or weakened this activity. Wild type MBP-OsGAP1 and MBP-only were included as positive and negative controls, respectively. Clu1, MBP-OsGAP1 with Cluster 1 mutated; Clu2, MBP-OsGAP1 with Cluster 2 mutated, etc. B, validation of the differential phospholipid-binding abilities of Cluster 1, 2, 3, and 4 mutants using membrane lipid strips (Echelon, P-6002).
Subsequently, we adopted the membrane lipid strips to examine the affinities of native and Cluster 1, 2, 3, and 4 mutants toward 15 biologically important membrane lipids. MBP showed a background affinity to two phospholipids, PtdIns 4-phosphate and PtdIns 4,5-bisphosphate. We confirmed that Cluster 2 and 4 mutants lost the affinities toward most of the membrane lipids, whereas Cluster 1 mutant harbored affinities similar to those of the wild type OsGAP1. In addition, the Cluster 3 mutant exhibited weakened or lost affinities toward some lipid species when compared with the wild type OsGAP1 protein (Fig. 3B). Based on the differential phospholipid binding abilities, mutants that are able (Cluster 1 mutant) or unable (Cluster 2 and 4 mutants) to bind phospholipids were employed for functional studies (see below).
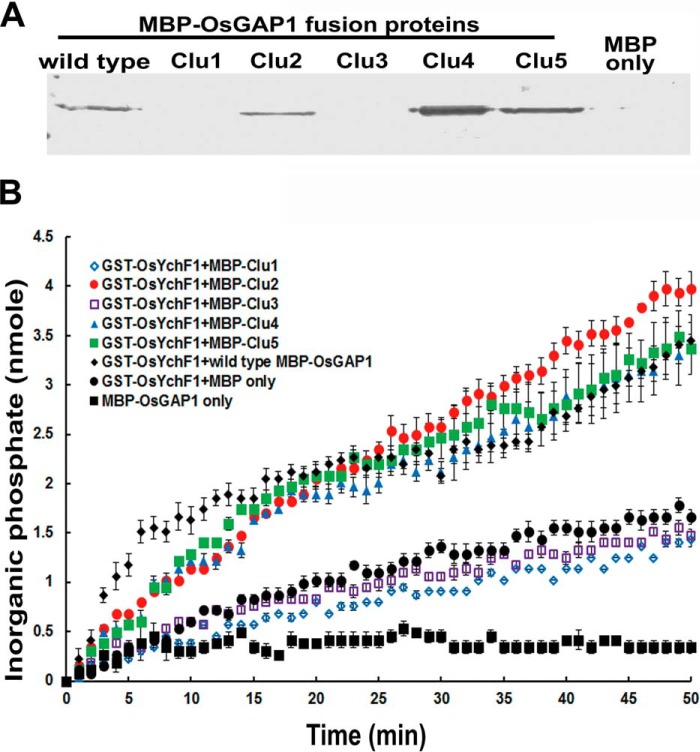
In vitro pull-downs of MBP-OsGAP1 fusion proteins with GST-OsYchF1 showed that mutations in Clusters 1 and 3, but not Cluster 2, 4, or 5, of OsGAP1 abolished the binding to OsYchF1 (Fig. 4A). Subsequently, the GTPase-stimulating activity of OsGAP1 on OsYchF1 was also lost in the mutants of Clusters 1 and 3 (Fig. 4B). Therefore, we postulated that Clusters 1 and 3 are located on the OsYchF1-binding surface, whereas Clusters 4, 5, 2, and 3 are located on the phospholipid-binding surface (Fig. 1C).
FIGURE 4.
Site-directed mutagenesis of OsGAP1 to dissect its binding to OsYchF1. Interaction of OsYchF1 with wild type and mutant OsGAP1 proteins. A, in vitro pull-down of the wild type and five mutant MBP-OsGAP1 fusion proteins by GST-OsYchF1-bound MagneGSTTM glutathione particles (MagneGSTTM protein purification system). Mutations in Clusters 1 and 3 abolished the binding of OsGAP1 to OsYchF1. Clu1, MBP-OsGAP1 with Cluster 1 mutated; Clu2, MBP-OsGAP1 with Cluster 2 mutated; etc. Anti-MBP antibodies were used for detection. MBP alone was used as a negative control. B, activation of the GTPase activity by wild type and mutant OsGAP1 proteins. GTPase activities were monitored by incubating the wild type and mutant MBP-OsGAP1 proteins with GST-OsYchF1 in the presence of GTP. Enzymatic activities were measured by the amount of inorganic phosphate released during GTP hydrolysis using the EnzChekTM phosphate assay kit (Molecular Probes). Mutations in Clusters 2, 4, and 5 in OsGAP1 did not affect the GTPase-activating activity on OsYchF1. As negative controls, MBP-OsGAP1 without OsYchF1 did not show any GTPase activities. MBP alone also did not stimulate the GTPase activity of OsYchF1. Error bars, S.E.
To perform functional studies, we selected mutants in Cluster 1 (loss of OsYchF1 binding) and Clusters 2 and 4 (loss of phospholipid binding) for further analyses. These mutants exhibited distinct changes in binding activities. Cluster 1 and Cluster 4 represent unique features of this class of C2 domains. Cluster 1 is responsible for the binding to an unconventional G protein (OsYchF1), which is a function unique to this group of C2-domain proteins, whereas Cluster 4 is located in the loop between the special βA and βB strands. Based on previous studies on other C2 domains (30, 34–37), the residues on Cluster 2 were predicted to be the ones most likely to contribute to phospholipid binding. Mutations in Cluster 2 therefore can be used as a negative control.
Binding to OsYchF1 Is Essential for the Functions of OsGAP1 in Both Defense and Abiotic Stress Responses
We previously set up a gain-of-function heterologous system using transgenic A. thaliana to show the protective effects of OsGAP1 under both pathogen attacks and abiotic stresses (11–13). Using the same system, we then tested the importance of the ability to bind OsYchF1 and phospholipids for the functions of OsGAP1.
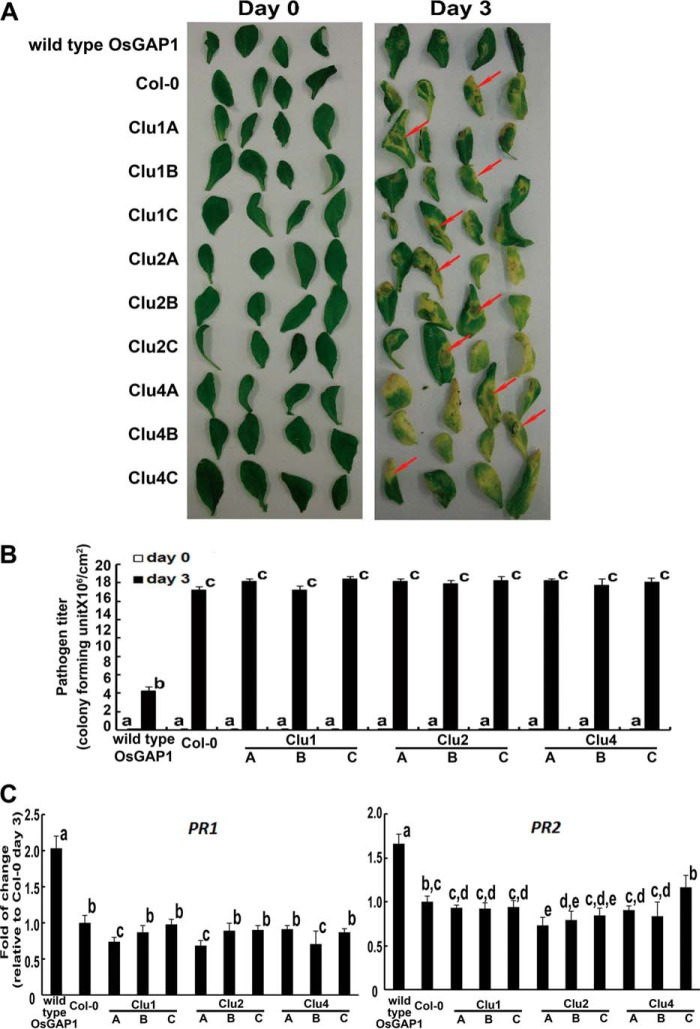
Mutations in Cluster 1, which caused a loss of OsYchF1 binding ability (and subsequently GTPase activation activity) of OsGAP1 (Fig. 4), provide a useful tool to study the physiological importance of OsYchF1 binding. When transgenic A. thaliana expressing the wild type OsGAP1 was inoculated with the pathogenic bacterium Pseudomonas syringae pv. tomato DC3000 (Pst DC3000), it displayed smaller lesion areas, lower pathogen titers, and higher expression levels of PR genes compared with the untransformed control (Col-0) (Fig. 5). This is consistent with our previous report (13). However, mutations in Cluster 1, which abolished the binding to OsYchF1, also eliminated the protective effect of OsGAP1 against Pst DC3000 (Fig. 5). The expression levels of different constructs in the transgenic lines under these conditions were found to be comparable (data not shown).
FIGURE 5.
Pathogen inoculation tests on transgenic A. thaliana expressing wild type and mutant OsGAP1s. A, disease symptoms developed by A. thaliana lines expressing the wild type or mutant OsGAP1 proteins (Clusters 1, 2, and 4) after inoculation with Pst DC3000. The symptoms were highlighted by red arrows. Transgenic lines expressing any of the mutant OsGAP1s displayed similar lesions to untransformed Col-0, whereas the wild type OsGAP1-transformed line was relatively unaffected by the pathogens. B, pathogen titer estimated 3 days after inoculation. Transforming Arabidopsis lines with any one of the mutant OsGAP1s did not offer any additional protection against pathogen invasion compared with the untransformed line (Col-0), whereas the wild type OsGAP1-transformed line had a significantly lower pathogen titer than all of the other lines. Error bars, S.E. of at least three samples. Three independent homozygous lines expressing each of the three mutant OsGAP1s were tested. Statistical analysis was performed using one-way analysis of variance combined with the Games-Howell post hoc test. Different lowercase letters above the bars represent statistically distinct groups at p < 0.05. Two biological repeats were performed, and similar results were obtained. C, the relative expression of defense marker genes (PR1 and PR2). RNA samples were taken 3 days after inoculation, and gene expression was estimated by real-time RT-PCR. Error bars, S.E. of at least three samples. Three independent homozygous lines expressing each of the three mutant OsGAP1s were tested. The transgenic line expressing the wild type OsGAP1 was included as the positive control. Col-0, untransformed wild type A. thaliana. The expression level in Col-0 was set to 1 for normalizing. Statistical analysis was performed using one-way analysis of variance combined with the Games-Howell post hoc test. Different lowercase letters above the bars represent statistically distinct groups at p < 0.05. Two biological repeats were performed, and similar results were obtained.
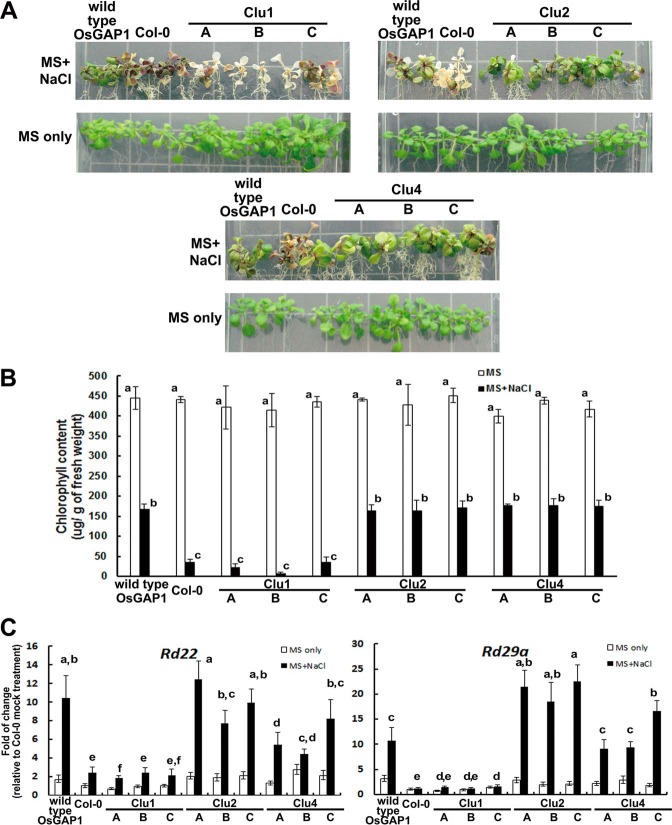
We also tested the protective functions of OsGAP1 against salinity stress by comparing transgenic A. thaliana expressing the wild type versus mutated OsGAP1 proteins. As expected, the wild type OsGAP1 could increase salinity stress tolerance (Fig. 6). The OsGAP1-transgenic plants maintained higher chlorophyll contents and expression levels of stress-adaptive genes, such as AtRD22 and AtRD29A (38–40). On the other hand, mutations in Cluster 1 abolished such protections (Fig. 6). These results demonstrated that the ability of OsGAP1 to bind OsYchF1 is critical for its functions in both defense and salt stress responses.
FIGURE 6.
Salt stress responses of transgenic A. thaliana expressing wild type and mutant OsGAP1s. A, representative phenotypes under salt stress. Phenotypes were observed after 10-day-old seedlings were transferred to MS agar supplemented with 150 mm NaCl for another 10 days. Salt treatment was applied to transgenic A. thaliana expressing either wild type or mutant OsGAP1s (Clu1, -2, and -4). B, the degree of chlorosis was quantified by chlorophyll measurement. Clu1 mutant-transformed and untransformed wild type A. thaliana were significantly more affected by salt stress than transgenic A. thaliana expressing wild type OsGAP1 and Clu2 and Clu4 mutants. C, the relative expression of salt-responsive genes (rd22 and rd29a). The RNA samples were taken 1 day after salt treatment. Three independent homozygous lines expressing each of the three mutant OsGAP1s were studied. The transgenic line expressing the wild type OsGAP1 was included as the positive control. Col-0, untransformed wild type A. thaliana. The expression in Col-0 grown on MS-only medium was set to 1 for reference. Statistical analysis was performed using one-way analysis of variance with the Games-Howell post hoc test. Different lowercase letters above each bar represent statistically distinct groups at p < 0.05. Two biological repeats were performed, and similar results were obtained.
Binding to Phospholipids Is Essential for the Function of OsGAP1 on Plant Defense but Not on Abiotic Stress Responses
Whereas OsGAP1 can bind directly to phospholipids, OsYchF1 was found to associate indirectly with phospholipids only in the presence of OsGAP1 (12). Mutants in Clusters 2 and 4 lost their ability to bind phospholipids but could still bind to OsYchF1 (Fig. 4). These constructs can be used to delineate the importance of phospholipid binding for the functions of OsGAP1.
Similar to the results with the mutant in Cluster 1, transgenic A. thaliana expressing mutants in Clusters 2 and 4 no longer displayed enhanced plant defense responses, different from those expressing the wild type OsGAP1 (Fig. 5). Because mutants in Clusters 2 and 4 could still bind to OsYchF1, the results indicated that in addition to binding to OsYchF1, the binding of OsGAP1 to phospholipids is also essential for it to enhance plant defense responses.
On the contrary, mutations in Clusters 2 and 4 did not exhibit obvious effects on the protective functions of OsGAP1 against salinity stress, whereas mutations in Cluster 1 resulted in similar responses to salt stress as the untransformed Col-0 (Fig. 6), indicating that OsGAP1 alleviates abiotic stress via a pathway associated with OsYchF1 but independent of phospholipid binding.
Discussion
In previous studies, we showed that OsGAP1 is involved in activating defense and abiotic stress responses (11–13). Specifically, OsGAP1 exhibits two important interactions essential for its physiological functions. First, it can bind to phospholipids (12). Second, it can bind to the unconventional G-protein OsYchF1 and stimulate its NTPase activity (12, 13). Here, we determined the crystal structure of OsGAP1. Based on the structural and sequence analyses, we have identified five clusters of conserved residues on the surface of OsGAP1. Our results showed that residues in Clusters 1 and 3 are involved in binding OsYchF1, whereas residues in Clusters 2–5 are involved in binding phospholipids (Figs. 1C and 3).
The binding of OsGAP1 to the unconventional G-protein OsYchF1 is a unique feature among C2-domain proteins, which we have also previously demonstrated in the Arabidopsis homologues AtGAP1/AtCAR1 and AtYchF1 (11). Through the phospholipid-binding ability of OsGAP1, OsYchF1 can be translocated from the cytosol to the plasma membrane after wounding (12). Such protein trafficking forms an early signal during pathogenic attacks (41). This explains why the ability to bind both phospholipids and OsYchF1 is required for OsGAP1 to stimulate plant defense responses (Fig. 5). Interestingly, other single-C2-domain proteins, such as AtCAR4 and OsPBP1, can also translocate between the cytoplasm and the plasma membrane (7, 10), suggesting that the C2 domain may play a role in controlling the subcellular localization of the protein, primarily by interacting with phospholipids reversibly. On the other hand, when the plant is subjected to abiotic stress, both OsGAP1 and OsYchF1 remain in the cytosol (11). Therefore, the ability of OsGAP1 to bind phospholipids is not critical for it to effect salinity stress responses (Fig. 6).
Besides OsGAP1, other single-C2-domain proteins are also found to be associated with both plant defense and abiotic stress responses. For example, AtBAP1 from A. thaliana suppresses plant defenses by inhibiting programmed cell death (an essential reaction of the hypersensitive response) (8, 9). On the other hand, AtBAP1 or its homologue AtBAP2 alleviates oxidative stress in yeast (9), suggesting a possibly generally positive role of plant C2-domain proteins in abiotic stress responses.
A recent report on Arabidopsis AtCAR4 (AtGAP1), a homologue of OsGAP1, suggested its involvement in the abscisic acid signaling pathway (10). Deletion of the extra β-hairpin and α-helix abolished the interactions between abscisic acid receptors and the plasma membrane. The alanine mutant of AtCAR4 at Asp-85 and Asp-87, which forms part of a conserved calcium-binding pocket, loses the calcium-dependent phospholipid binding ability, yet the extra domain and the calcium binding pocket may not be the only sites for phosopholipid or protein interaction. Therefore, not all of the possible interacting surfaces were identified in AtCAR4 (AtGAP1). Also, the connection between lipid specificity and the physiological role of the phospholipid interaction of AtCAR4 (AtGAP1) was not discussed explicitly. Our findings here give additional important information on the molecular mechanism of this group of plant-specific single-C2-domain proteins.
Because YchF1 homologues are highly conserved among different groups of living organisms (12) and AtYchF1 exhibits very similar properties to OsYchF1, including the ability to bind both OsGAP1 and AtGAP1/AtCAR4 (11), we could make use of transgenic A. thaliana in functional assays to examine the consequences of losing the ability of OsGAP1 to bind either OsYchF1 or phospholipids.
There are other reports showing the regulation of G proteins by C2-domain proteins in both animals and plants. For example, the C2-domain protein RGS in Caenorhabditis elegans can directly associate with the Gα subunits of heterotrimeric G proteins (42), whereas the C2 domain is also present in an effector molecule that binds to the human Rab8a GTPase to regulate membrane trafficking (43). In addition, C2 domains have been found in several GAPs, including SynGAP (44), RasGAPs (45), RhoGAPs (46), and ArfGAPs (47, 48).
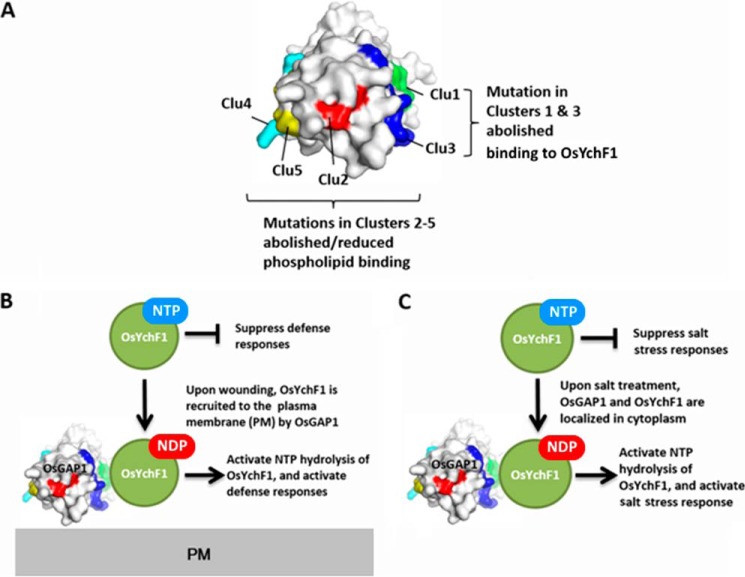
Based on the current data and our previous findings (11–13), we put forth a working model to explain the structure-function relationship of OsGAP1 (Fig. 7). Residues in Clusters 4, 5, 2, and 3 are involved in phospholipid binding, whereas residues in Clusters 1 and 3 are essential for interaction with OsYchF1. OsYchF1 is found to be a negative regulator of both plant defense responses and abiotic stress responses (11–13). Upon wounding, OsGAP1 recruits OsYchF1 to the plasma membrane by interacting with phospholipids and activates the NTPase activities of OsYchF1 (12). Defense responses are activated (12). Under salt stress, OsGAP1 and OsYchF1 are both localized in the cytosol and OsGAP1 activates the NTPase activities of OsYchF1. Salt stress responses are activated (11).
FIGURE 7.
Working model to explain the structure-function relationship of OsGAP1. A, the structure of OsGAP1 is shown in a surface representation. Mutations in Clusters 1 (green) and 3 (blue) abolished the interaction with OsYchF1, whereas mutations in Clusters 2 (red), 3, 4 (cyan), and 5 (yellow) abolished/reduced the phospholipid binding of OsGAP1. B, the interaction of OsGAP1 and OsYchF1 on the plasma membrane is involved in plant defense responses. C, the interaction of OsGAP1 and OsYchF1 in the cytosol is involved in salt stress responses.
In summary, we identified two functional surfaces in OsGAP1 through structure-function analyses. Binding to OsYchF1 is always essential for OsGAP1 to function in both defense and abiotic stress responses, whereas interaction with phospholipids is only important in defense responses. These findings will advance our understanding of this group of novel C2-domain proteins and their modes of function. They also shed light on the regulatory mechanisms of the understudied YchF-type unconventional G-proteins.
Author Contributions
H.-M. L., Y.-L. Y., M.-Y. C., R. M., K.-B. W., and M.-L. C. designed the experiments. Y.-L. Y., M.-Y. C., K.-P. L., and M.-H. Y. performed site-directed mutagenesis and related functional tests. R. M., Y.-H. F., and K.-B. W. produced the crystal and analyzed the data of x-ray crystallography. H.-M. L., M.-Y. C., and Y.-L. Y. wrote the manuscript. All authors proofread and revised the manuscript.
Acknowledgments
We thank Iris Tong for providing technical help and Jee-Yan Chu for copy-editing this manuscript.
This work was supported by grants from the Hong Kong Research Grants Council (General Research Fund: 468013; Area of Excellence: AoE/M-05/12) (to H. M. L., K. B. W., and M. L. C.). The authors declare that they have no conflicts of interest with the contents of this article.
- GAP
- GTPase-activating protein
- MBP
- maltose-binding protein
- PtdIns
- phosphatidylinositol.
References
- 1. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 2. Meijer H. J. G., Munnik T. (2003) Phospholipid-based signaling in plants. Annu. Rev. Plant Biol. 54, 265–306 [DOI] [PubMed] [Google Scholar]
- 3. Stahelin R. V. (2009) Lipid binding domains: more than simple lipid effectors. J. Lipid Res. 50, S229–S304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang D., Aravind L. (2010) Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene 469, 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cho W., Stahelin R. V. (2006) Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta 1761, 838–849 [DOI] [PubMed] [Google Scholar]
- 6. Kopka J., Pical C., Hetherington A. M., Müller-Röber B. (1998) Ca2+/phospholipid-binding (C2) domain in multiple plant proteins: novel components of the calcium-sensing apparatus. Plant Mol. Biol. 36, 627–637 [DOI] [PubMed] [Google Scholar]
- 7. Yang W. Q., Lai Y., Li M.-N., Xu W.-Y., Xue Y.-B. (2008) A novel C2-domain phospholipid-binding protein, OsPBP1, is required for pollen fertility in rice. Mol. Plant 1, 770–785 [DOI] [PubMed] [Google Scholar]
- 8. Yang H., Li Y., Hua J. (2006) The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J. 48, 238–248 [DOI] [PubMed] [Google Scholar]
- 9. Yang H., Yang S., Li Y., Hua J. (2007) The Arabidopsis BAP1 and BAP2 genes are general inhibitors of programmed cell death. Plant Physiol. 145, 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodriguez L., Gonzalez-Guzman M., Diaz M., Rodrigues A., Izquierdo-Garcia A. C., Peirats-Llobet M., Fernandez M. A., Antoni R., Fernandez D., Marquez J. A., Mulet J. M., Albert A., Rodriguez P. L. (2014) C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis. Plant Cell 26, 4802–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheung M.-Y., Li M.-W., Yung Y.-L., Wen C.-Q., Lam H.-M. (2013) The unconventional P-loop NTPase OsYchF1 and its regulator OsGAP1 play opposite roles in salinity stress tolerance. Plant Cell Environ. 36, 2008–2020 [DOI] [PubMed] [Google Scholar]
- 12. Cheung M.-Y., Xue Y., Zhou L., Li M.-W., Sun S. S.-M., Lam H.-M. (2010) An ancient P-Loop GTPase in rice is regulated by a higher plant-specific regulatory protein. J. Biol. Chem. 285, 37359–37369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung M.-Y., Zeng N. Y., Tong S.-W., Li W.-Y. F., Xue Y., Zhao K. J., Wang C. L., Zhang Q., Fu Y. P., Sun Z., Sun S. S.-M., Lam H.-M. (2008) Constitutive expression of a rice GTPase-activating protein induces defense responses. New Phytologist 179, 530–545 [DOI] [PubMed] [Google Scholar]
- 14. Fong Y. H., Wong H. C., Chuck C. P., Chen Y. W., Sun H., Wong K. B. (2011) Assembly of preactivation complex for urease maturation in Helicobacter pylori: crystal structure of UreF-UreH protein complex. J. Biol. Chem. 286, 43241–43249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 16. Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 1022–1025 [DOI] [PubMed] [Google Scholar]
- 17. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 19. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 20. Kapust R. B., Waugh D. S. (1999) Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brears T., Liu C., Knight T. J., Coruzzi G. M. (1993) Ectopic overexpression of asparagine synthetase in transgenic tobacco. Plant Physiol. 103, 1285–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bechtold N., Pelletier G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266 [DOI] [PubMed] [Google Scholar]
- 23. Koncz C., Schell J. (1986) The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396 [Google Scholar]
- 24. Katagiri F., Thilmony R., He S. Y. (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. in The Arabidopsis Book (Last R., Chang C., Jander G., Kliebenstein D., McClung R., Millar H., eds) p. e0039, American Society of Plant Biologists, Rockville, MD: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moran R., Porath D. (1980) Chlorophyll determination in intact tissues using N,N-dimethylformamide. Plant Physiol. 65, 478–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moran R. (1982) Formulae for determination of chlorophyllous pigments extracted with N,N-dimethylformamide. Plant Physiol. 69, 1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Webb M. R. (1992) A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. U.S.A. 89, 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nalefski E. A., Falke J. J. (1996) The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 5, 2375–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ochoa W. F., Garcia-Garcia J., Fita I., Corbalan-Garcia S., Verdaguer N., Gomez-Fernandez J. C. (2001) Structure of the C2 domain from novel protein kinase Ce: a membrane binding model for Ca2+-independent C2 domains. J. Mol. Biol. 311, 837–849 [DOI] [PubMed] [Google Scholar]
- 30. Guerrero-Valero M., Ferrer-Orta C., Querol-Audí J., Marin-Vicente C., Fita I., Gómez-Fernández J. C., Verdaguer N., Corbalán-García S. (2009) Structural and mechanistic insights into the association of PKCa-C2 domain to PtdIns(4,5)P2. Proc. Natl. Acad. Sci. U.S.A. 106, 6603–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Davletov B. A., Südhof T. C. (1993) A single C2 domain from synaptotagmin-I is sufficient for high-affinity Ca2+/phospholipid binding. J. Biol. Chem. 268, 26386–26390 [PubMed] [Google Scholar]
- 32. Stevens C. F., Sullivan J. M. (2003) The synaptotagmin C2A domain is part of the calcium sensor controlling fast synaptic transmission. Neuron 39, 299–308 [DOI] [PubMed] [Google Scholar]
- 33. Kelly S. M., Price N. C. (2000) The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Peptide Sci. 1, 349–384 [DOI] [PubMed] [Google Scholar]
- 34. Bittova L., Sumandea M., Cho W. (1999) A structure-function study of the C2 domain of cytosolic phospholipase A2: identification of essential calcium ligands and hydrophobic membrane binding residues. J. Biol. Chem. 274, 9665–9672 [DOI] [PubMed] [Google Scholar]
- 35. Fukuda M., Kojima T., Mikoshiba K. (1996) Phospholipid composition dependence of Ca2+-dependent phospholipid binding to the C2A domain of synaptotagmin IV. J. Biol. Chem. 271, 8430–8434 [DOI] [PubMed] [Google Scholar]
- 36. Kojima T., Fukuda M., Aruga J., Mikoshiba K. (1996) Calcium-dependent phospholipid binding to the C2A domain of a ubiquitous form of double C2 protein (Doc2 β). J. Biochem. 120, 671–676 [DOI] [PubMed] [Google Scholar]
- 37. Sutton R. B., Sprang S. R. (1998) Structure of the protein kinase C beta phospholipid-binding C2 domain complexed with Ca2+. Structure 6, 1395–1405 [DOI] [PubMed] [Google Scholar]
- 38. Msanne J., Lin J., Stone J. M., Awada T. (2011) Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234, 97–107 [DOI] [PubMed] [Google Scholar]
- 39. Shinozaki K., Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227 [DOI] [PubMed] [Google Scholar]
- 40. Wang H., Zhou L., Fu Y., Cheung M.-Y., Wong F. L., Phang T.-H., Sun Z., Lam H.-M. (2012) Expression of an apoplast-localized BURP-domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress. Plant Cell Environ. 35, 1932–1947 [DOI] [PubMed] [Google Scholar]
- 41. Leon J., Rojo E., Sanchez-Serrano J. J. (2001) Wound signalling in plants. J. Exp. Bot. 52, 1–9 [DOI] [PubMed] [Google Scholar]
- 42. Sato M., Moroi K., Nishiyama M., Zhou J., Usui H., Kasuya Y., Fukuda M., Kohara Y., Komuro I., Kimura S. (2003) Characterization of a novel C. elegans RGS protein with a C2 domain: evidence for direct association between C2 domain and Gαq suunit. Life Sci. 73, 917–932 [DOI] [PubMed] [Google Scholar]
- 43. Hokanson D. E., Bretscher A. P. (2012) EPI64 interacts with Slp1/JFC1 to coordinate Rab8a and Arf6 membrane trafficking. Mol. Biol. Cell 23, 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pena V., Hothorn M., Eberth A., Kaschau N., Parret A., Gremer L., Bonneau F., Ahmadian M. R., Scheffzek K. (2008) The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. EMBO Rep. 9, 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gawler D. J., Zhang L. J. W., Moran M. F. (1995) Mutation-deletion analysis within p120 GAP, a GTPase-activating protein for p21 ras. Biochem. J. 307, 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hallam S. J., Goncharov A., McEwen J., Baran R., Jin Y. (2002) SYD-1, a presynaptic protein with PDZ, C2 and rhoGAP-like domains, specifies axon identity in C. elegans. Nat. Neurosci. 5, 1137–1146 [DOI] [PubMed] [Google Scholar]
- 47. Jensen R. B., Lykke-Andersen K., Frandsen G. I., Nielsen H. B., Haseloff J., Jespersen H. M., Mundy J., Skriver K. (2000) Promiscuous and specific phospholipid binding by domains in ZAC, a membrane-associated Arabidopsis protein with an ARF GAP zinc finger and a C2 domain. Plant Mol. Biol. 44, 799–814 [DOI] [PubMed] [Google Scholar]
- 48. Zhuang X., Xu Y., Chong K., Lan L., Xue Y., Xu Z. (2005) OsAGAP, an ARF-GAP from rice, regulates root development mediated by auxin in Arabidopsis. Plant Cell Environ. 28, 147–156 [DOI] [PubMed] [Google Scholar]