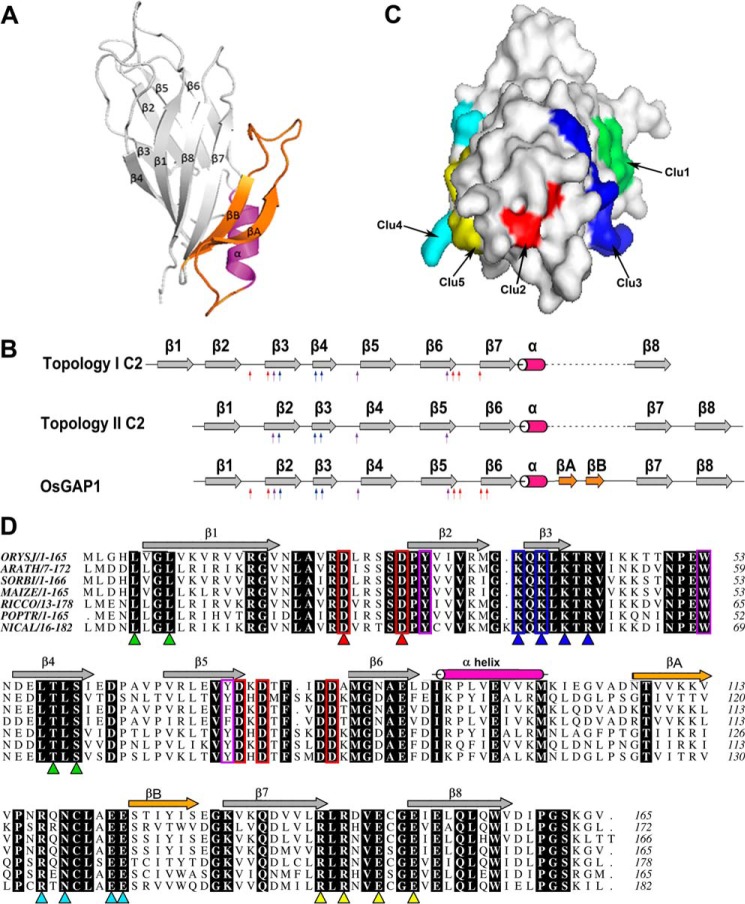
FIGURE 1.
Structural analysis of OsGAP1. A, a ribbon diagram showing the structure of OsGAP1 as resolved by x-ray crystallography to a resolution of 1.63 Å. B, a schematic diagram showing that OsGAP1 exhibits overall a type II C2-domain topology but possesses signature residues conserved among type-I C2 domains. Red arrows indicate conserved aspartate residues in the putative lipid-binding loop typical of type I topology. Blue and purple arrows, positively charged residues in the putative cationic β-groove and conserved aromatic residues, respectively (conserved in both topologies). Two unique β-strands newly discovered only in OsGAP1 (βA and βB) are indicated in orange. C, a surface diagram showing the locations of the five clusters of mutated residues. Clusters 1 and 3 (Clu1 and Clu3) are closer in one face, whereas Clusters 4, 5, 2, and 3 (Clu4, Clu5, Clu2, and Clu3) line up at another angle. D, sequence alignment of plant homologues of OsGAP1. Identical residues among homologues are highlighted in black. Five clusters of amino acids designated for mutagenesis are indicated by triangles (green, Cluster 1; red, Cluster 2; blue, Cluster 3; cyan, Cluster 4; yellow, Cluster 5). The main criteria for choosing these residues were that they must be surface residues and must be conserved among other plant GAP1 homologues. Conserved residues common to the PKC-C2 domain are boxed (red, aspartate residues making up a calcium-binding pocket; blue, lysine patch; purple, aromatic residues). Five clusters of residues were chosen as the targeted mutation sites. ORYSJ, homologue from O. sativa subsp. Japonica (GenBankTM accession number NP_001046709); it is the same gene encoding OsGAP1. ARATH, homologue from A. thaliana (GenBankTM accession number NP_188425); it is the same gene encoding AtCAR4. SORBI, homologue from Sorghum bicolor (GenBankTM accession no. XP_002455977). MAIZE, homologue from Zea mays (GenBankTM accession number NP_001132168). RICCO, homologue from Ricinus communis (GenBankTM accession number XP_002530486). POPTR, homologue from Populus trichocarpa (GenBankTM accession number XP_006383322). NICAL, homologue from Nicotiana alata (GenBankTM accession number ACD40010).