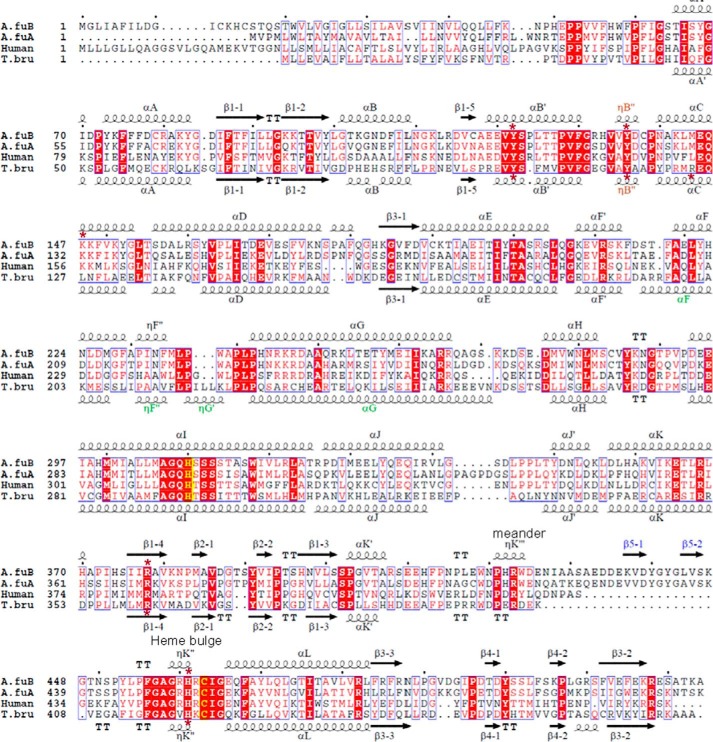
FIGURE 2.
Sequence alignment of CYP51 proteins from A. fumigatus (A.fuB and A.fuA), human, and a protozoan pathogen, T. brucei (T.bru). The alignment was generated in ClustalW and processed in ESPript to add secondary structure information on A.fuB (top) and T.bru (bottom), using molecules A of Protein Data Bank files 4UYL and 3GW9, respectively. The amino acid sequence identity between A.fuB and A.fuA is 59%. Within the Aspergillus genus, CYP51B identities range from 78% (Aspergillus niger) to 99% (Aspergillus fischerianus), whereas CYP51A identities range from 69% (Aspergillus nidulans) to 96% (A. fischerianus) (not shown). The identities between A.fuB versus human and A.fuB versus T.bru CYP51 amino acids are 33 and 23%, respectively. The alignment shows that, regardless of low amino acid sequence identity, the length and location of the secondary structural elements in A.fuB and T.bru CYP51s match very well, except for the FG arm segment, which is longer in T.bru (in green), and the additional β-bundle in A.fuB (strands β5-1 and β5-2, in blue); this segment appears to be specific to fungal CYP51 and so far has not been seen in the structures of CYPs from any other families. The CYP51-specific helical turn in the SRS1 region (ηB″) is shown in brown. The heme-coordinated cysteine and the conserved CYP51-specific histidine (proton delivery, helix I) are colored in yellow; the five residues bonded with porphyrin propionates are marked with asterisks.