Fig. 3.
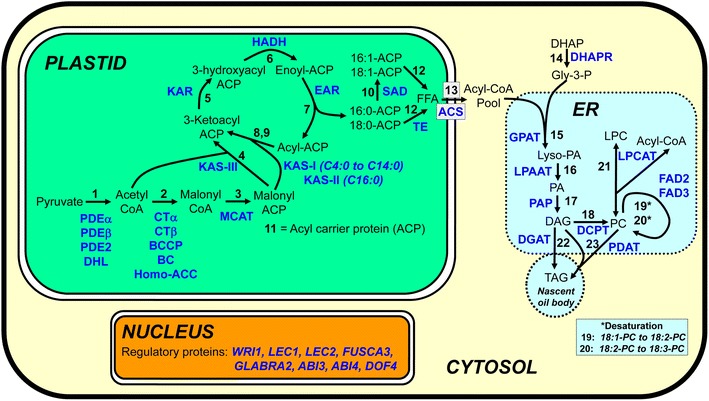
Summary of mapped candidate genes involved in the biosynthesis of storage lipids in J. curcas. The genes, indicated in blue text, are as follows: Plastid—(1) PDEα α-subunit of the pyruvate dehydrogenase (PDH) complex, PDEβ β-subunit of the PDH complex, PDE2 dihydrolipoyl transacetylase component of the PDH complex and PDE3 dihydrolipoamide dehydrogenase component of the PDH complex; (2) CTα α-subunit of the heteromeric acetyl-coA carboxylase (ACCase) complex, CTβ β-subunit of the heteromeric ACCase complex, BCCP biotin carboxyl carrier protein and BC biotin-carboxylase subunit of the heteromeric ACCase complex; (3) MCAT malonyl-CoA:ACP malonyltransferase (4,8 and 9) and KAS 3-ketoacyl-ACP synthase; (5) KAR 3-ketoacyl-ACP reductase; (6) HADH 3-hydroxylacyl-ACP dehydratase; (7) EAR enoyl-ACP reductase; (10) SAR stearoyl-ACP reductase; (11) ACP acyl carrier protein; (12) ACP-TE acyl-ACP thioesterase; (13) ACS acyl-CoA synthetase. Cytosol —(14) DHAPR dihydroxyacetone phosphate reductase. Endoplasmic Reticulum (ER)—(15) GPAT glycerol-3-phosphate acyltransferase; (16) LPAAT lysophosphatidic acid acyltransferase; (17) PAP phosphatidate phosphatase; (18) DCPT diacylglycerol:choline phosphatidyltransferase; (19, 20) FAD fatty acid desaturase; (21) LPCAT 1-acylglycerol-3-phosphocholine acyltransferase; (22) DGAT diacylglycerol acyltransferase; (23) PDAT phospholipid:diacylglycerol acyltransferase. Nucleus —regulatory proteins including Wrinkled1 (WRI1), Leafy Cotyledon 1 & 2 (LEC1 & LEC2), FUSCA3, GLABRA2, Abscisic Acid Insensitive 3 & 4 (ABI3 & ABI4) and DOF4. Abbreviations used for pathway intermediates (black) include DHAP dihydroxyacetone phosphatase, Gly-3-P glycerol-3-phosphate, Lyso-PA lysophosphatidic acid, PA phosphatidic acid, DAG diacylglycerol, TAG triacylglycerol, PC phosphatidylcholine and LPC lysophosphatidylcholine
