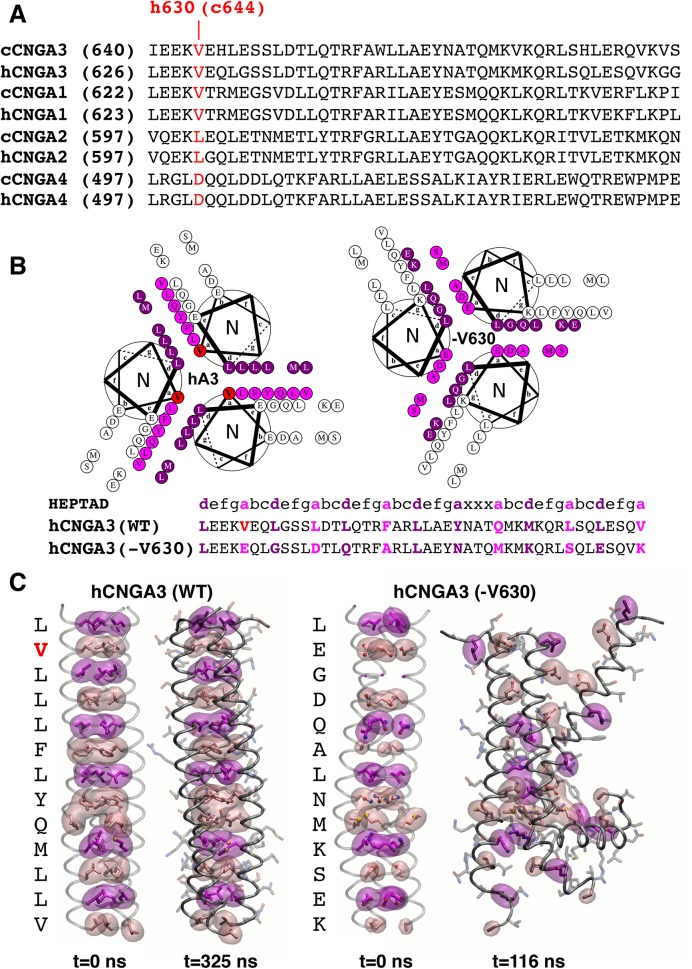
Fig 4. The V644 deletion alters CLZ core structure indicating unfolding of coiled-coil complex.
(A) Sequence alignment of the CLZ (C-terminal leucine zipper) domains in CNGA-type subunits and conservation of the V644 residue. Note that canine V644 corresponds to V630 of human CNGA3. c = canine; h = human. (B) Helical wheel diagrams looking down the superhelical axis of the CLZ coiled-coil from the N- to C- terminus. Heptad a positions are marked in pink, d positions are denoted in magenta, and V630 is shown in red. The V630del shifts residues from b and e heptad positions to the core a and d positions and causes the predominantly hydrophobic residues of the coiled-coil core in the wild-type structure (left) to shift destabilizing charged and small residues in the mutant complex (right). Sequence diagram of the wild-type and V630del mutant heptad is shown below. (C) Molecular dynamics trajectories and models of the wild-type and mutant CLZ trimeric structures. Snapshots before (t = 0 ns) and after (t = 325 ns) simulation for the wild-type CNGA3-CLZ (left). The CLZ core clearly remains intact with small perturbations near the solvent exposed N- and C- termini over long timescales. Snapshots before (t = 0 ns) and after (t = 116 ns) simulation for the V630del mutant model (right). The V630 deletion dramatically alters the previously stabilizing residue-residue interactions, leading to disruption of the CLZ coiled-coil and helical structure. Small red and blue spheres correspond to oxygen and nitrogen atoms, respectively. See also S4and S5 Figs for the full-length molecular dynamics simulations.