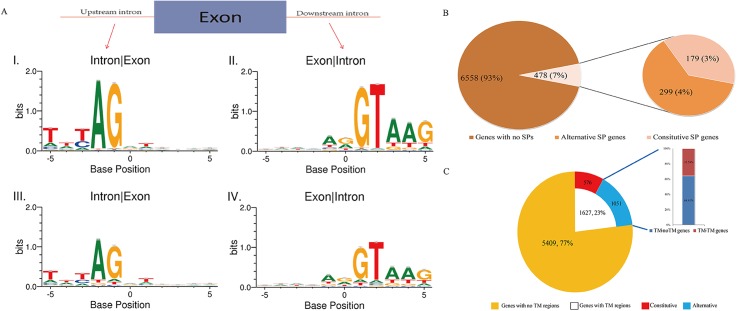
Fig 4. Binding motif search and alternative splicing in excretory and secretory proteins encoding genes.
A, Sequence situations in boundaries of Cassette exons and alternative exons. Four bases frequencies in both ends of an exon and flank introns in five positions were calculated and showed by Weblogo 3.3 (http://weblogo.threeplusone.com/). I, left boundaries of cassette exons, namely intron-exon boundaries. II, right boundaries of cassette exons, namely exon-intron boundaries. III, left boundaries of alternative exons. IV, right boundaries of cassette exons. B, Alternative splicing in genes encoding proteins with signal peptides (SPs). Signal peptides were predicted using online tool SignalP 4.1 server, setting as default (D-cutoff for SignalP-noTM networks is 0.45 and D-cutoff for SignalP-TM networks is 0.5). Numbers in the pie chart imply gene amounts and percentages. Genes with no SPs: genes predicted as encoding transcripts with no signal peptides. Alternative SP genes: genes predicted as encoding multiple transcripts with or without signal peptides. Constitutive SP genes: genes predicted as encoding multiple transcripts with signal peptides. C, Alternative splicing in genes encoding proteins with transmembrane (TM) domains. Transmembrane domains were predicted using online tool TMHMM server2.0, setting as default. Annotations in the pie chart indicate gene amount and percentage. Constitutive: genes encode transcripts possessing transmembrane domains without alternative splicing found. Alternative: genes encode transcripts possessing transmembrane domains with alternative splicing found; TM- noTM genes: genes encode multiple transcripts with and without TM domains; TM-TM genes: genes encode multiple transcripts possessing different amount of TM domains.