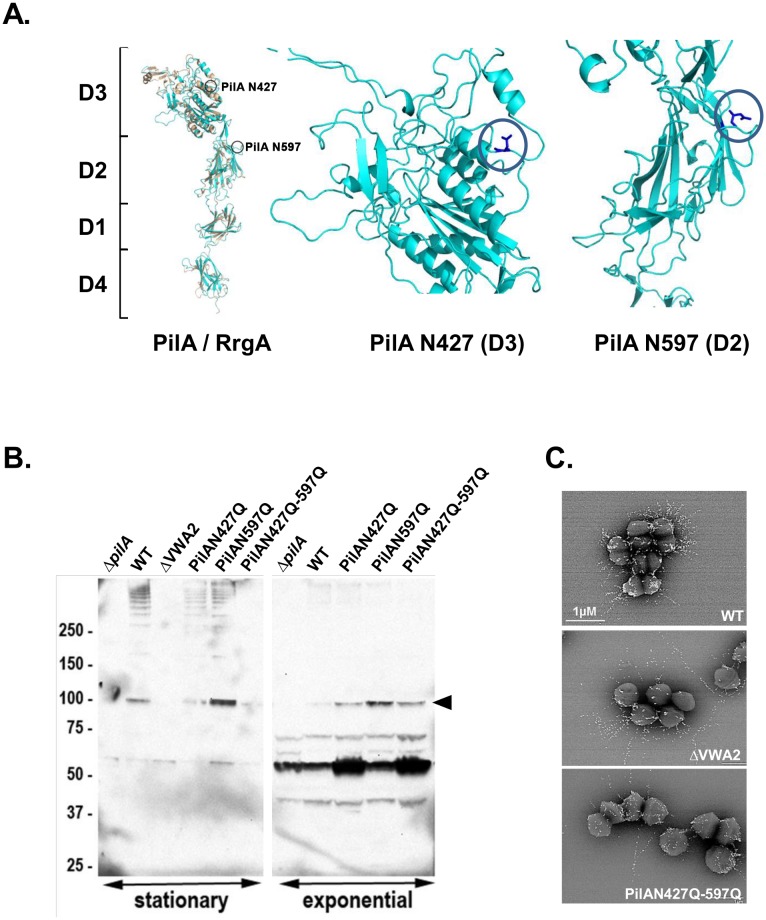
Fig 4. Role of putative N-glycosylated asparagyl residues in PilA stability.
(A) Structural modeling of PilA (blue ribbon) compared to the S. pneumoniae homolog RrgA (orange ribbon). Domain organization (D1 to D4) appears to be conserved, position of in silico putative predicted N-glycosylated aspargyl residues N427 and N597 in domains D3 and D2 are indicated (blue circle). (B) Proteins anchored to the cell-wall were isolated from S. agalactiae strain NEM316 and its isogenic pilA mutants harvested at both exponential and stationary growth phase, separated on 4%–12% Criterion XT SDS-PAGE gel, and detected by immunoblotting with specific anti-PilA antiserum. Equivalent amount corresponding to 500 μl of the initial culture was loaded in each well. The PilA monomer is indicated by a black arrow. The high-molecular-weight species correspond to PilA polymers while the lower band at 50 kDa is most likely a degradation product. (C) Immuno-electron-microscopy (IEM) analyses of the pilus subunits PilB. S. agalactiae wild-type strain NEM316 (WT) and its isogenic pilus mutants (ΔVWA2, and PilAN427Q-N597Q) were incubated with rabbit polyclonal antibody raised against PilB. Antibodies were conjugated to 10 nm gold particles.