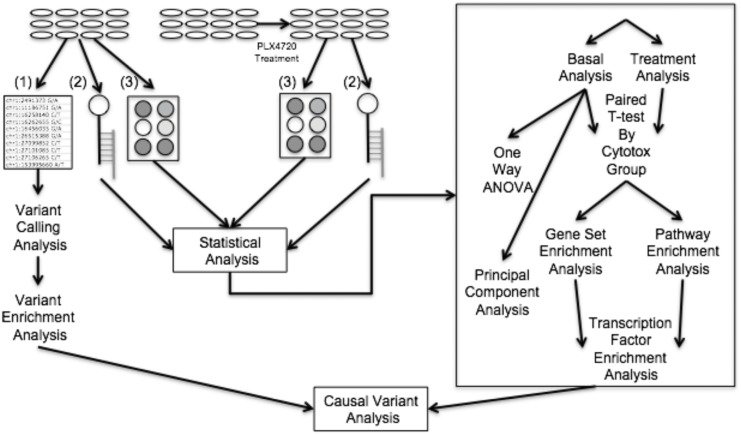
Fig 1. Functional genomic data generated and analysis workflow.
Flow-chart including transcriptome, proteome and exome data generated and integrative analysis of these data sets. 12 cell lines (not drug-treated) were characterized using exome sequencing (1), gene expression arrays (2), and reverse phase protein arrays (3). Variant calling and enriched gene variants were identified by cross referencing our results with mutated genes associated with cancer identified by Lawrence et al [20]. Cell lines were exposed to PLX4720 and their responses were assayed with gene expression arrays and reverse phase protein arrays. Protein response to treatment was correlated with cytotoxic effects of PLX4720 treatment to identify proteins that might be mitigating the cytotoxic response. Cytotoxicity (Cytotox) groups were identified by clustering the cytotoxicity data. Differential gene expression responses to treatment within each cytotoxicity group were identified and underwent both gene set and pathway enrichment analysis via MSigDB and Pathway Express respectively. Pathway enrichment analysis revealed ErbB signaling as a key response to treatment and gene set enrichment analysis revealed a number of transcription factors that are enriched which putatively regulate ErbB signaling pathway genes.