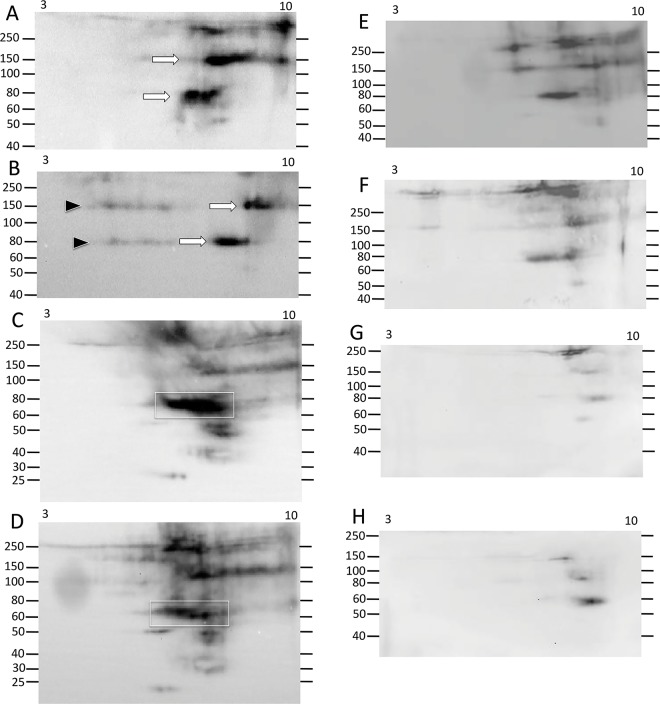
Fig 9. A wide range of changes in the MW/pI of Ni-Sp185/333 proteins can occur over time and in response to multiple challenges with different types of microbes.
(A) Prior to immune challenge, sea urchin 107 shows two major trains of Ni-Sp185/333 proteins of ~80 and ~150 kDa (arrows). (B) After the first challenge with V. diazotrophicus, the majority of the proteins in the trains shift to more basic (arrows), with an expansion of acidic proteins within the same trains (black arrowheads). (C) Prior to challenge with Bacillus sp, sea urchin 107 shows a wide repertoire of Sp185/333 proteins, particularly those of ~60 to 80 kDa and pI of ~7 to 8 (white box). (D) After challenge with Bacillus sp, there is a slight decrease in the intensity of the Ni-Sp185/333 proteins, particularly those of ~60 to 80 kDa/pI ~6 to 8 (white box). (E) The second challenge with Bacillus sp results in a decrease in the intensity of the repertoire of Sp185/333 proteins. (F) The third challenge with Bacillus sp further decreases the repertoire and intensity of the Ni-Sp185/333 proteins. (G) Two weeks after challenge with Bacillus sp the Ni-Sp185/333 protein repertoire shows a further decrease in the number of spots and their intensity. (H) Subsequent challenge with V. diazotrophicus does not induce an increase in the Ni-Sp185/333 protein repertoire, but shows further decreases in diversity. Images (A, B, E-H) were cropped at the bottom because they do not show spots of less than 40 kDa.