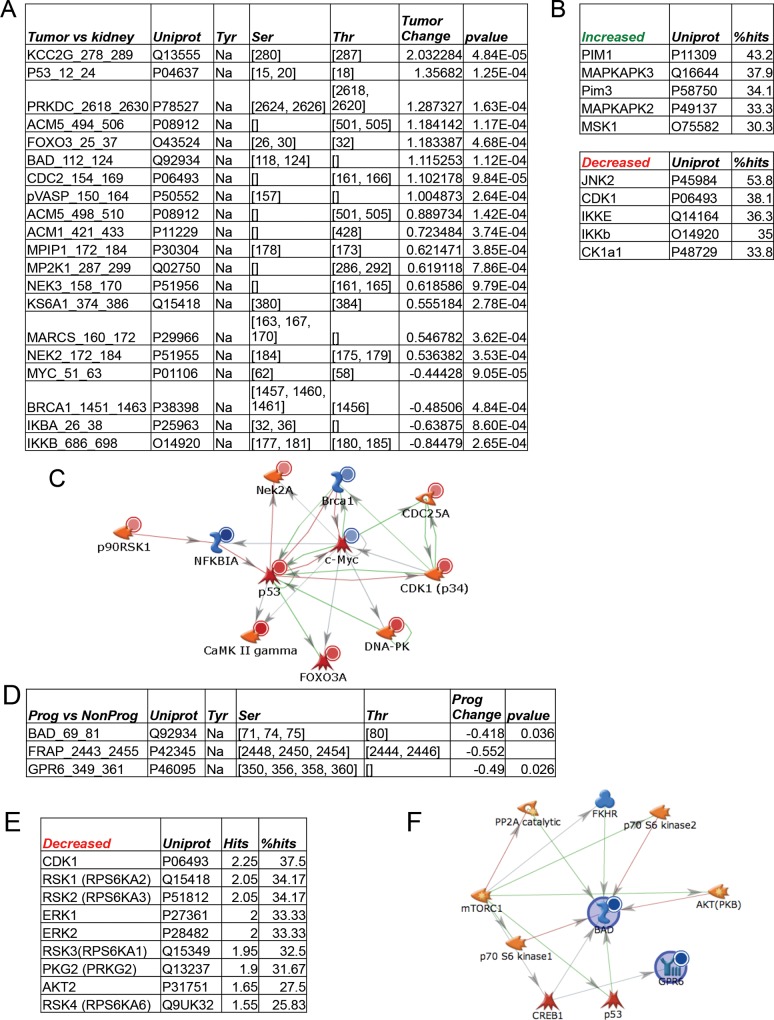
Fig 4. Kinases altered in CC-RCC and relationship to clinical outcome.
CC-RCC tumors that had matched normal fresh frozen material available (n = 12) were directly compared and statistically different phosphopeptides (p<0.001) were identified and are shown in (A). These significant peptides were used to query Kinexus Phosphonet as in Fig 3 (and as described in Materials and Methods). Predicted upstream kinases that distinguish CC-RCC from matched normal kidney (indicated as increased or decreased in CC-RCC relative to normal kidney) are shown in (B). GeneGo MetaCore Network Modeling of the proteins that contain the significantly altered phosphopeptides (Listed as Uniprot ID’s in A) is shown in (C). Red circles indicate increased phosphorylation of the peptide while blue circles indicate decreased substrate phosphorylation. A supervised analysis of the CC-RCC tumors was performed to determine kinomic differences between patients who remained locally controlled after a minimum follow up of 18 months (NonProg) and those who progressed (Prog). Peptides significantly altered between these groups (D) were used to query Kinexus Phosphonet as above and are shown in (E) which were decreased. GeneGo MetaCore Network Modeling of the proteins containing the significantly altered phosphopeptides is shown in (F) where blue circles indicate decreased phosphorylation of the peptide.