Abstract
M cells in follicle-associated epithelium (FAE) covering intestinal lymphoid follicles serve as a portal of entry for particulate antigens (Kanaya and Ohno, 2014 [1]). Despite their biological significance, molecular mechanisms that govern M-cell differentiation and function have not been fully elucidated. MicroRNAs (miRNAs) have a role to control host gene expression profiles that modulate cellular physiology and characteristic. Many studies have shown that miRNAs regulate diverse biological processes including developmental timing, differentiation and growth control of cells and tissues (Ivey and Srivastava, 2010 [2]). miRNAs are also relevant to differentiation and function of intestinal epithelium (McKenna et al., 2010 [3]; Runtsch et al., 2014 [4]). Expression profiles and functions of miRNAs in the intestinal epithelium have been examined in jejunal and colonic mucosa [3]. In contrast, those in FAE remain uncharacterized. To address this deficiency, we isolated Peyer's Patch (PP) FAE and villous epithelium (VE) surrounding the FAE, and compared the miRNA expression profiles of FAE and VE by microarray analysis. This revealed that 43 miRNAs were up-regulated, whereas 9 miRNAs were down-regulated, in FAE compared to VE. A unique pattern of miRNA expression by FAE may reflect important diversity in cellular phenotypes and/or functional features of FAE. All microarray data has been deposited at GEO under accession number GSE46264.
Keywords: MicroRNA, Peyer's patch, Follicle-associated epithelium, Villus epithelium, M cell
| Specifications | |
|---|---|
| Organism/cell line/tissue | Mus musculus |
| Sex | Female |
| Sequencer or array type | Agilent Scanner (G2505B) |
| Data format | Analyzed |
| Experimental factors | Healthy mice in SPF facility |
| Experimental features | Conducted miRNA expression profiling of intestinal epithelium in BALB/cA mice |
| Consent | N/A |
| Sample source location | Yokohama, Japan |
1. Direct link to deposited data
2. Experimental design, materials and methods
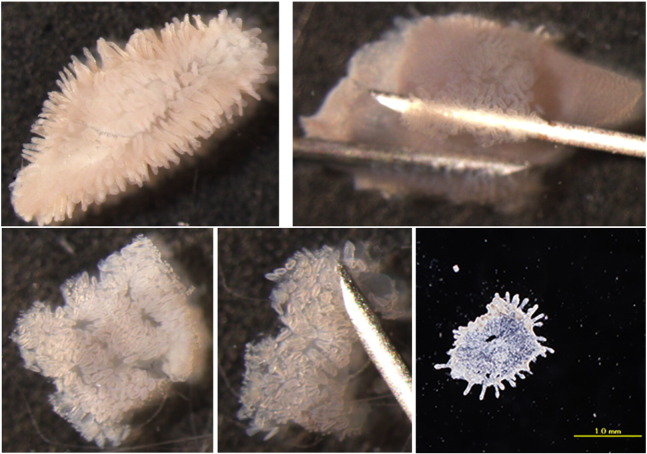
PP FAE and VE were isolated from 10 week old SPF BALB/cA mice. Briefly, intestinal tissue was soaked in Hank's balanced salt solution containing 30 mM EDTA for 20 min at 4 °C. After incubation, FAE and VE were isolated by dissection with 26G needles using stereomicroscopic monitoring, as shown in Fig. 1 (Hase et al., 2005 [5]). Isolated epithelial cell sheets were maintained in ice-cold HBSS before RNA extraction. Total RNA was extracted from isolated FAE and VE using mirVana kit (Ambion) according to the manufacturer's instructions. miRNA expression profile was assessed by using Mouse miRNA Microarray Release 12.0 (Agilent Technologies). A hundred nanograms of total RNA was labeled with Agilent miRNA Complete Labeling Reagent and Hyb Kits (Agilent) according to the manufacturer's instructions. The labeled RNA was used to probe the array plates via hybridization for 20 h at 55 °C. Array plates were subsequently washed and then scanned with an Agilent G2565CA Microarray Scanner (Agilent Technologies). Data analysis was performed with Genespring software version 11.0 (Agilent Technologies). Two-fold expression difference between FAE and VE were considered to be significant.
Fig. 1.
Procedure of FAE and VE isolation from Peyer's patch.
Scatter plot of normalized miRNA expression profiles showed that 43 miRNAs were up-regulated, and nine miRNAs were down-regulated, in FAE compared with VE by at least two-fold (Fig. 2). To efficiently promote particulate antigen uptake, the cellular composition and functions of FAE differ dramatically from VE. VE is dominated by absorptive enterocytes, with scattered goblet cells and enteroendocrine cells. By contrast, FAE contains few goblet or enteroendocrine cells and instead is characterized by the presence of antigen-sampling M cells [1]. Reflecting the difference in epithelial cell compositions, gene expression is differently regulated between FAE and VE [5]. Consistent with this, our microarray data showed that miRNA profiles between FAE and VE were different (See Table 1, Table 2). A unique pattern of miRNA expression for FAE may reflect important cellular phenotypes as well as functional features in FAE. We believe that detailed analysis of FAE/M cell-specific miRNAs will likely provide new insights related to mechanisms of M-cell development and function.
Fig. 2.
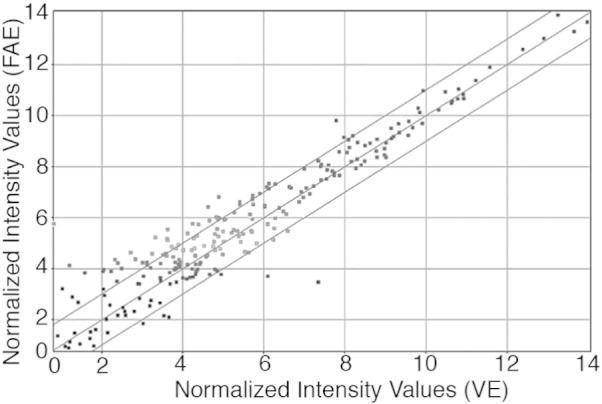
Comparison of miRNA expression level between FAE and VE. Lines indicate the threshold of two-Fold change.
Table 1.
miRNAs upregulated more than 2-fold in the FAE as compared to VE.
| miRNA name | miRBase accession number | Fold change |
|---|---|---|
| mmu-miR-15a* | MIMAT0004624 | 7.28 |
| mmu-miR-511 | MIMAT0004940 | 5.69 |
| mmu-miR-872* | MIMAT0004935 | 5.16 |
| mmu-miR-582-3p | MIMAT0005292 | 4.51 |
| mmu-miR-193 | MIMAT0000223 | 4.23 |
| mmu-miR-466d-3p | MIMAT0004931 | 3.95 |
| mmu-miR-365 | MIMAT0000711 | 3.66 |
| mmu-miR-204 | MIMAT0000237 | 3.47 |
| mmu-miR-149 | MIMAT0000159 | 3.32 |
| mmu-miR-335-5p | MIMAT0000766 | 3.29 |
| mmu-miR-210 | MIMAT0000658 | 3.22 |
| mmu-miR-18a* | MIMAT0004626 | 3.21 |
| mmu-miR-485* | MIMAT0003129 | 3.08 |
| mmu-miR-466 h | MIMAT0004884 | 3.01 |
| mmu-miR-452 | MIMAT0001637 | 2.97 |
| mmu-miR-191* | MIMAT0004542 | 2.87 |
| mmu-miR-290-5p | MIMAT0000366 | 2.84 |
| mmu-miR-1224 | MIMAT0005460 | 2.70 |
| mmu-miR-34a | MIMAT0000542 | 2.69 |
| mmu-miR-466a-3p | MIMAT0002107 | 2.69 |
| mmu-miR-335-3p | MIMAT0004704 | 2.68 |
| mmu-miR-1894-5p | MIMAT0007877 | 2.67 |
| mmu-miR-150 | MIMAT0000160 | 2.62 |
| mmu-miR-297a* | MIMAT0004864 | 2.61 |
| mmu-miR-203* | MIMAT0004547 | 2.54 |
| mmu-miR-764-5p | MIMAT0003894 | 2.50 |
| mmu-miR-328 | MIMAT0000565 | 2.43 |
| mmu-miR-296-5p | MIMAT0000374 | 2.42 |
| mmu-miR-669a | MIMAT0003477 | 2.42 |
| mmu-miR-206 | MIMAT0000239 | 2.37 |
| mmu-miR-326 | MIMAT0000559 | 2.36 |
| mmu-miR-346 | MIMAT0000597 | 2.33 |
| mmu-miR-207 | MIMAT0000240 | 2.29 |
| mmu-miR-211 | MIMAT0000668 | 2.25 |
| mmu-miR-1196 | MIMAT0005857 | 2.22 |
| mmu-miR-467 g | MIMAT0005854 | 2.20 |
| mmu-miR-467e* | MIMAT0005294 | 2.18 |
| mmu-miR-376b | MIMAT0001092 | 2.17 |
| mmu-miR-376a | MIMAT0000740 | 2.16 |
| mmu-miR-669 h-3p | MIMAT0005842 | 2.13 |
| mmu-miR-294 | MIMAT0000372 | 2.09 |
| mmu-miR-672 | MIMAT0003735 | 2.07 |
| mmu-miR-34b-3p | MIMAT0004581 | 2.06 |
Table 2.
miRNAs upregulated more than twice in the VE compared to the FAE.
| miRNA name | miRBase accession number | Fold change |
|---|---|---|
| mmu-miR-143 | MIMAT0000247 | 16.31 |
| mmu-miR-451 | MIMAT0001632 | 5.79 |
| mmu-miR-654-3p | MIMAT0004898 | 5.76 |
| mmu-miR-224 | MIMAT0000671 | 2.92 |
| mmu-miR-10a | MIMAT0000648 | 2.34 |
| mmu-miR-219 | MIMAT0000664 | 2.17 |
| mmu-miR-101a | MIMAT0000133 | 2.17 |
| mmu-miR-494 | MIMAT0003182 | 2.07 |
| mmu-let-7e | MIMAT0000524 | 2.06 |
Acknowledgments
This study was supported in part by Grant-in-Aid for Research Activity Start-up (G.N.) (Grant number: 22890238), Grants-in-Aid for Young Scientists (B) (G.N.) (Grant number: 24790485) and Young Scientists (A) (K.H.) (Grant number: 24249029), Scientific Research (A) (H.O.) (Grant number: 22689017) from the Japan Society for the Promotion of Science, Grant-in-Aid For Scientific Research on Priority Areas (K.H. and H.O.) (Grant number: 19041072) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Advanced Research and Development Programs for Medical Innovation from the Japan Agency for Medical Research and Development (H.O.) (Grant number: 15gm0710009h0002), and Sasakawa Scientific Research Grant from the Japan Science Society (G.N.) (Grant number:22-453).
References
- 1.Kanaya T., Ohno H. The Mechanisms of M-cell Differentiation. Biosci. Microbiota Food Health. 2014;33:91–97. doi: 10.12938/bmfh.33.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivey K.N., Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 3.McKenna L.B., Schug J., Vourekas A., McKenna J.B., Bramswig N.C., Friedman J.R., Kaestner K.H. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654–1664. doi: 10.1053/j.gastro.2010.07.040. 1664 e1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Runtsch M.C., Round J.L., O'Connell R.M. MicroRNAs and the regulation of intestinal homeostasis. Front. Genet. 2014;5:347. doi: 10.3389/fgene.2014.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hase K., Ohshima S., Kawano K., Hashimoto N., Matsumoto K., Saito H., Ohno H. Distinct gene expression profiles characterize cellular phenotypes of follicle-associated epithelium and M cells. DNA Res. 2005;12:127–137. doi: 10.1093/dnares/12.2.127. [DOI] [PubMed] [Google Scholar]