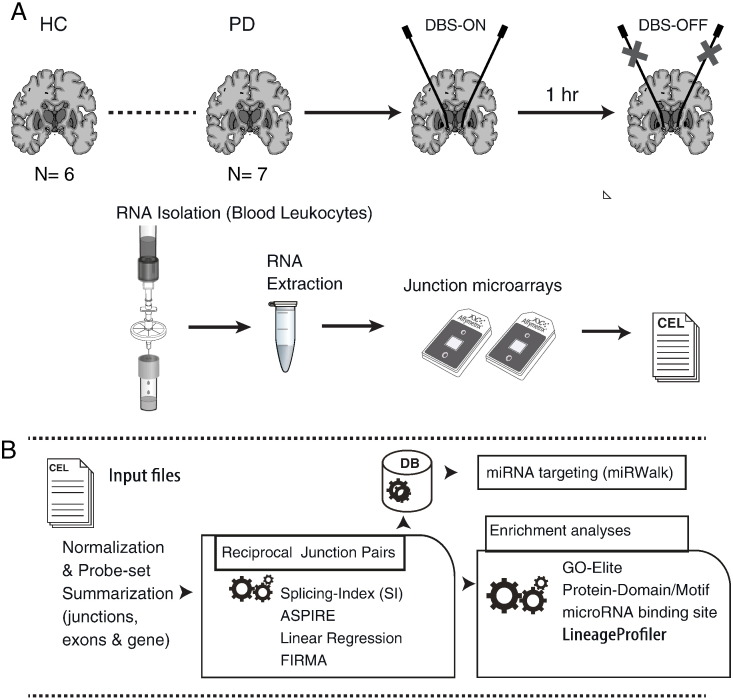
Fig. 2.
Experimental design and general analysis flow. (A) RNA was prepared from filtered blood leukocytes which were isolated from whole blood samples of PD patients a day prior to DBS, upon hospitalization from the PD patient participants, and post-DBS, upon clinical stabilization of symptoms following the electrical stimulation, both while being on stimulation (DBS-ON) and following 1 h off electrical stimulation (DBS-OFF). Age- and gender-matched (male) healthy volunteers served as a control group (HC). From each RNA sample, cDNA library was prepared (using the Affymetrix exon microarrays sample preparation protocol) and applied to Affymetrix junction prototype microarrays (HJAYs). (B) The junction array datasets were analyzed using AltAnalyze by applying several measures for alternative splicing, including a linear regression approach. Additionally, enrichment for microRNA (miRNA) binding sites and protein domains in the spliced regions was calculated, and functional analysis was conducted using the GO-Elite module of AltAnalyze. Cellular composition was computed using the software LineageProfiler module. Potential binding by miRNAs was also assessed using the miRWalk repository, for characterization of binding in 3′, 5′ and coding regions of targets that were detected as alternatively spliced.