Figure 7. Conservation of oRG Marker Gene Expression in Primates.
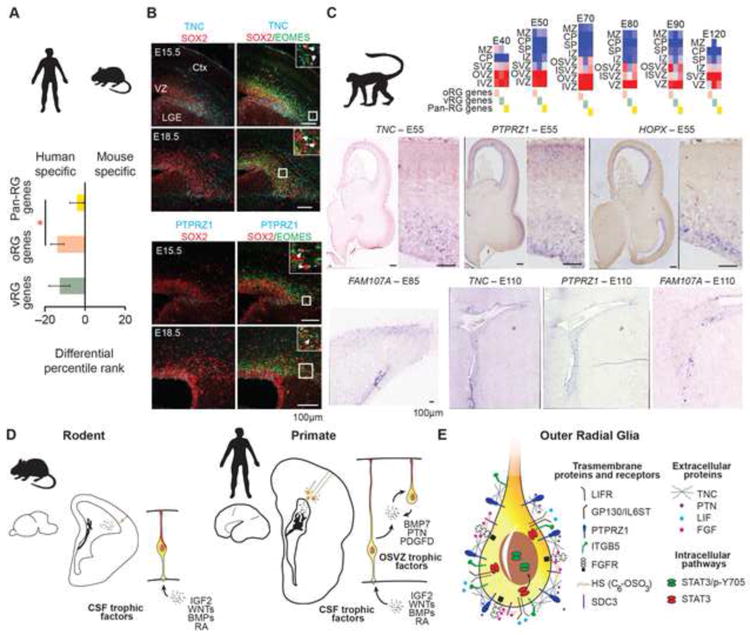
(A) Bar graph indicates average differential percentile rank of pan-RG, oRG, and vRG genes for radial glia gene coexpression network specificity in human and mouse. Compared with pan-RG genes (n=29), human oRG genes (n=66) show reduced conservation with mouse radial glia signature (p < 0.05, Wilcoxon rank sum test). (B) Mouse E15.5 and E18.5 cortical sections immunoreacted for Tnc and Ptprz1 along with Sox2 and Eomes. Inset images show a magnified view of SVZ region and arrows highlight examples of Sox2–positive, Eomes–negative cells that co-label for Tnc or Ptprz1. LGE- lateral ganglionic eminence, Ctx-cortex. (C) Heatmaps show average expression level of oRG, vRG and pan-RG genes in distinct regions of developing macaque cortex. IVZ – inner VZ, OVZ – outer VZ; The NIH Blueprint Non-Human Primate (NHP) Atlas. In situ hybridization of macaque cortex showing expression of oRG marker genes mirrors human trajectory. (D) Radial glia in small rodent brains are concentrated along the ventricle and access cerebrospinal fluid trophic factors directly via apical processes. In contrast, large primate brains contain numerous radial glia in the OSVZ. Local production of growth factors by radial glia may provide additional trophic support to oRG cells in the OSVZ niche. (E) Increased expression of extracellular matrix proteins that potentiate growth factor signaling and activation of LIFR/STAT3 pathway may further maintain stemness in oRG cells. See also Figure S7.
