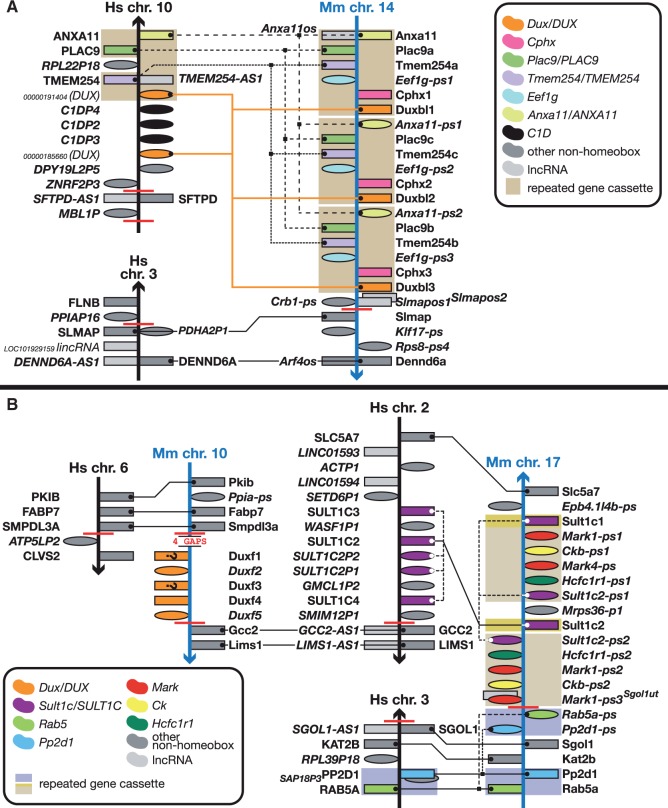
Figure 3.
Different Duxbl and DUX clusters in mouse and human and a mouse-specific Duxf cluster. Figure is not to scale. See figure for a guide to colours, Figure 1 for a guide to symbols and Figure 1 legend for notes on naming. (A) Mouse has seen an expansion of a gene cassette containing a Dux gene. Where mouse has three copies of the cassette, human only has one copy of each of the genes (where orthologues exist). This region is close to a synteny breakpoint. (B) A small cluster of five Duxf (pseudo)genes on mouse chromosome 10 has no equivalent in the human genome. For the genes marked with a question mark, it is unclear at this juncture whether these are the indicated biotypes as there is insufficient or conflicting evidence for an accurate determination of their biotype: coding genes could be pseudogenes and vice versa. The cluster is flanked by gaps and synteny breakpoints. Note the presence of a SULT1C cluster next to the human orthologue of Gcc2, the gene flanking the mouse Dux cluster. The mouse orthologue of this cluster has been subject to duplication and rearrangement as part of a six-gene cassette. Coincidentally, there is a Sult2a cluster next to the Obox cluster (Figure 1). There are many synteny breakpoints in these regions, indicating evolutionary instability.