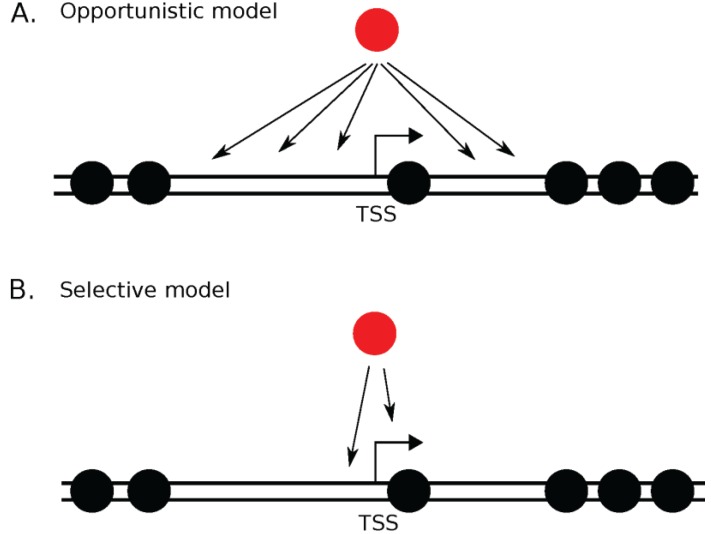
Figure 1.
Are histone variants opportunistic occupiers or do they have exclusive targets? Replication-dependent histone variants (canonical H3.1, H3.2, H4, H2A, and H2B) are assembled into chromatin during S phase. On the other hand, replication-independent histone variants are assembled throughout the cell cycle (or late M/early G1 for CENP-A). Without the replication fork guiding the site of assembly, why do histone variants localize to sites where they are found? Two models exist. In the opportunistic model (A) histone variants are deposited in the chromatin once a nucleosome free region is presented, irrespective of the underlying DNA sequence or chromatin context. In the selective model (B) histone variants are deposited in either a sequence-specific manner, (such as a TATA box) via pre-bound transcription factors or associated transcription machinery, or through its chaperone. TSS = Transcription Start Site.