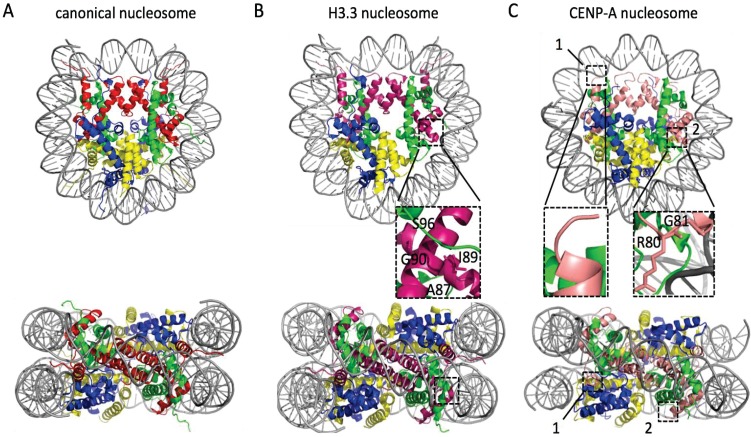
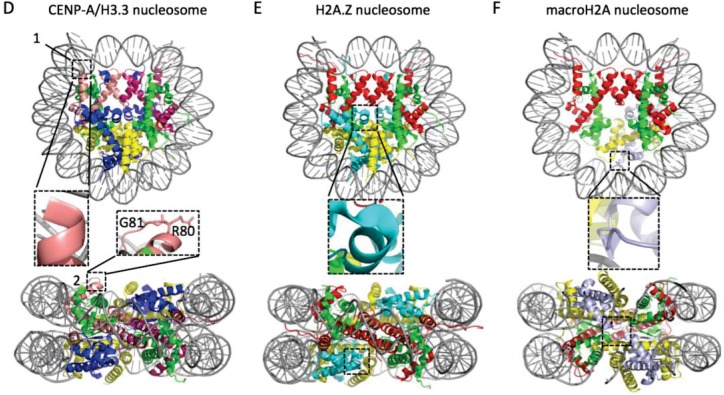
Figure 2.
Crystal structures of nucleosomes containing histone variants. In each panel the nucleosome is shown in top and side view. In general, the colors used are red for H3, green for H4, blue for H2A, yellow for H2B, and grey for DNA. For the nucleosomes with histone variants (B–F) boxed areas and corresponding insets magnify specific differences between the canonical nucleosome (A) and the respective variant nucleosome. In (B) pink marks H3.3, a variant that differ from the canonical histone H3.1 by five amino acids, four of which are in alpha helix 2 of the histone fold domain and directly interact with H4 (box). These residues allow the CAF1, and HIRA and DAXX chaperones to discriminate between H3.1 and H3.3, respectively. To highlight these differences, one pair of H2A/H2B was peeled away from the top view. In (C) the centromere-specific CENP-A (in salmon) nucleosome is shown. This nucleosome wraps ~121 bp compared to ~147 in all other nucleosomes. This is due to a shorter alpha N helix (box 1 and 1') and a longer loop 1 (box 2 and 2'). In (D) the heterotypic CENP-A/H3.3 nucleosome is shown using the respective colors for CENP-A and H3.3. This heterotypic nucleosome displays the independent structural characteristics unique to the two histone variants, where box D1 and D2 correspond to C1 and C2 respectively, and wraps DNA with a bimodal length distribution: 133 bp or canonical 147 bp. In (E) the structure of H2A.Z nucleosome is shown, where H2A.Z is colored cyan. Despite only having 60% sequence similarity to the canonical H2A, H2A.Z nucleosome is structurally almost identical to (A) and wraps ~147 bp. The extended acidic patch of H2A.Z, that is thought to be important for its unique functions (see text), is highlighted by a box. Finally, in (F), the macroH2A nucleosome is shown. Because the linker between the histone fold domain and the macro domain of macroH2A is too flexible, it could not be crystallized. Therefore, the macro domain and the histone fold domain were crystallized separately and here only the histone fold domain is shown. To highlight the flexible and hydrophobic loop1-loop1 interface with a box, one dimer of macroH2A/H2B is peeled away in the top view. The crystal structures were obtained from the RCSB Protein Data Bank using the following identification codes: (A) 1AOI; (B) 3AV2; (C) 3AN2; (D) 3WTP; (E) 1F66; and (F) 1U35 and visualized using PyMOL software version 1.7.6.0 (Schrödinger, Cambridge, MD, USA, 2015).