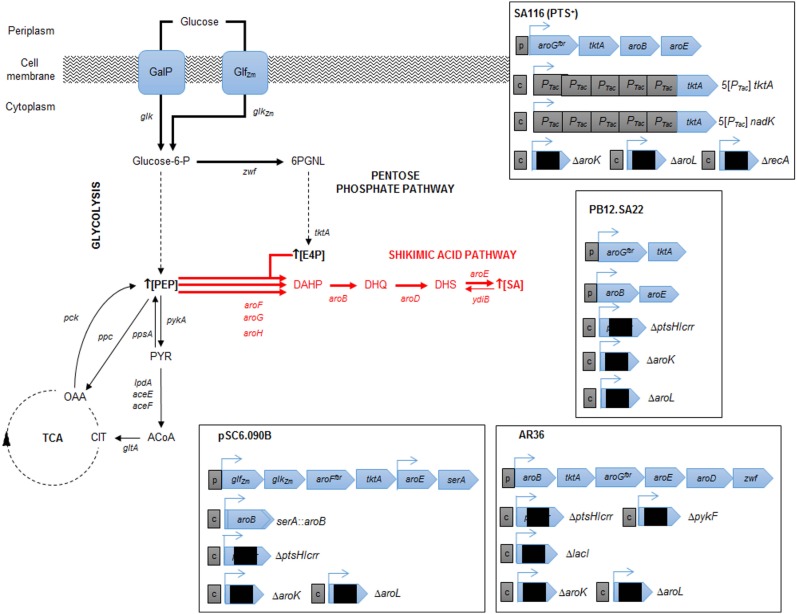
Figure 3.
Relevant engineered E. coli strains for SA production. Metabolic traits of E. coli derivative strains engineered for SA production resulting in highest SA titer and yield from glucose. The figure illustrates alterative glucose transporter GalP (galactose permease) selected by the cell after laboratory adaptive evolution process of a PTS− mutant (Flores et al., 1996, 2007; Aguilar et al., 2012). Glf (glucose facilitator) and Glk (glucokinase) from Z. mobilis (plasmid cloned). Resultant characteristics of engineered strains are shown for pSC6.090B (Chandran et al., 2003), PB12.SA22 (Escalante et al., 2010), AR36 (Rodriguez et al., 2013), and SA116 strain (Cui et al., 2014). CCM key intermediates and protein encoding genes: TCA, tricarboxylic acid pathway; E4P, erythrose-4-P; PGNL, 6-phospho d-glucono-1,5-lactone; PEP, phosphoenolpyruvate; PYR, pyruvate; ACoA, acetyl-CoA; CIT, citrate; OAA, oxaloacetate; zwf, glucose 6-phosphate-1-dehydrogenase; tktA, transketolase I; pykA, pyruvate kinase II; lpdA, aceE and aceF, coding for PYR dehydrogenase subunits; gltA, citrate synthase; pck, PEP carboxykinase; ppc PEP carboxylase; ppsA, PEP synthetase. SA pathway intermediates and genes: DAHP, 3-deoxy-d-arabino-heptulosonate-7-phosphate; DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; SA, shikimic acid. Continuous arrows indicate single enzymatic reactions; dashed arrows show several enzymatic reactions. Bold arrows show improved carbon flux. Black squares in plasmids/operons indicate gene interruption; c, chromosomal gene interruption or integration; p, plasmid-cloned genes.