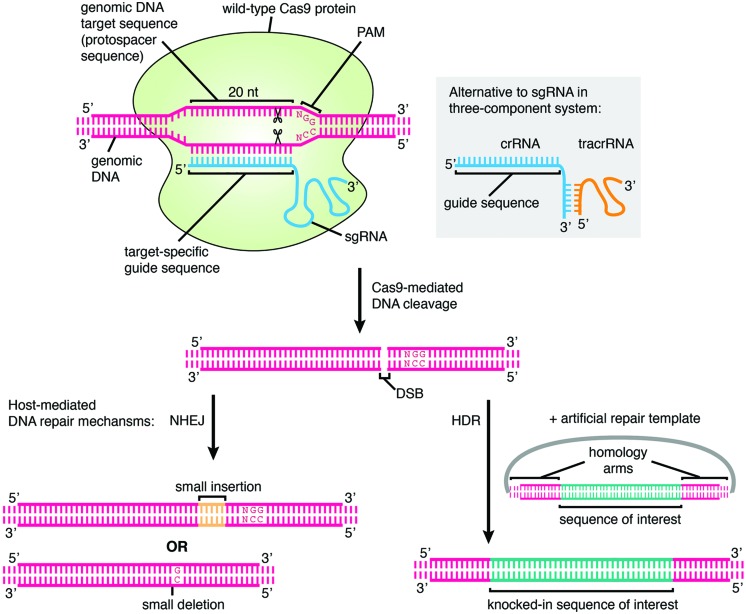
FIGURE 1.
The mechanism of genome editing using CRISPR/Cas9. The genomic DNA target, which must lie adjacent to a protospacer adjacent motif (PAM), is specified by a 20 nt user-generated guide sequence in the sgRNA or crRNA. The Streptococcus pyogenes PAM is shown. In the cell nucleus, Cas9 protein associates with the sgRNA or crRNA/tracrRNA and binds to the target sequence, cleaving both strands of the DNA at the site 3 nt upstream of the PAM. Cleavage results in a DSB which is repaired by host-mediated DNA repair mechanisms. In the absence of a repair template, error-prone NHEJ occurs which may lead to the formation of random short indels and thus frameshift mutations and disruption of gene function, and this represents the main method of CRISPR-mediated gene knockout generation. If an artificial repair template is provided, for example on a plasmid containing a sequence of interest flanked by homology arms, then HDR may occur, leading to the introduction of an exogenous DNA sequence at a specified genomic location. This is the basis for performing gene knock-in, tagging, and precise pre-specified insertions or deletions using CRISPR. If catalytically inactive Cas9 is used instead of wild-type Cas9, then the protein simply binds to the target locus and does not cleave the DNA.