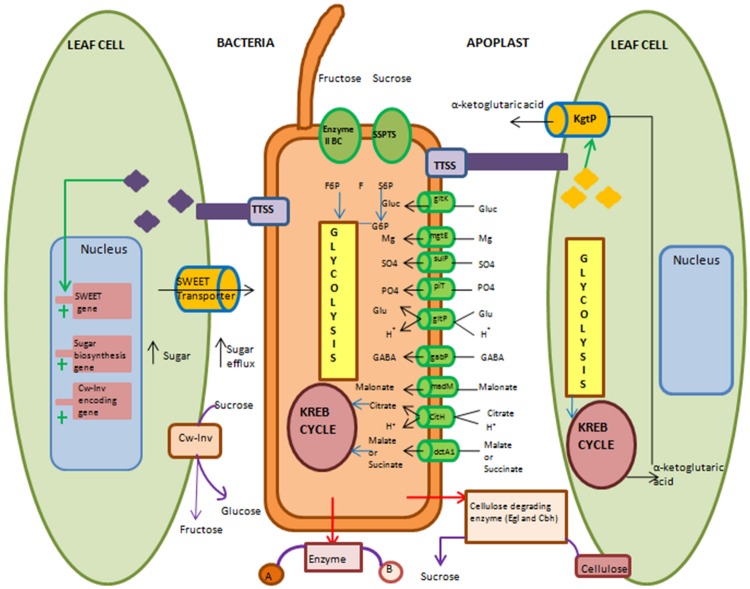
FIGURE 2.
Overview of molecular and biochemical events used by biotrophic or hemibiotrophic bacteria to acquire nutrients during apoplast colonization. Pathogenic bacteria use several strategies to acquire nutrients. They can either modulate their own machinery or manipulate plant cell machinery to acquire nutrients. During modulation of their own machinery, bacteria activate various transporters and take up nutrients that are present in apoplast. They can secrete cellulose degrading enzyme to release cell wall-bound nutrients (Cw-Inv). For uptake of less preferred nutrients they use two different ways. First, by secreting enzyme in apoplasm that converts undesirable form of nutrient into desirable form and then take up that desirable form of nutrient by transporter. Second, the uptake of less preferred nutrient by specific transporter and then suitably metabolize its energy. Sucrose specific phosphotransferase system is shown here for uptake of sucrose in the form of sucrose-6-phosphate (S6P) and catabolize it into fructose (F) and glucose-6-phosphate (G6P). Type III secretion system (TTSS) delivered effectors target expression of sugar transporter-encoding genes of host cell. Effector-mediated induction of SWEET (sugar will eventually be exported transporter) transporter for increasing sugar efflux in apoplast is shown here. Also induction of sugar biosynthesis genes for high sugar synthesis in cytosol and its movement to apoplast is shown. Expression of host cell wall-invertase-mediated conversion of sucrose into glucose and fructose in apoplast is depicted. α-Ketoglutaric acid transporter (KgtP) secreted by bacterial pathogen though TTSS and its localization in host cell membrane and the efflux of α-ketoglutaric acid from host cell into apoplasm is illustrated. Enzyme II BC, Fructose specific phosphotransferase system; SSPTS, Sucrose-specific phosphotransferase system; gltK, glucose ATP-binding cassette transporters (ABC) transporter permease; mgtE, magnesium transporter; fecC, Iron ABC transporter permease; sulP, sulphatepermease; piT, inorganic phosphate transporter; gltP, proton/glutamate symport; gabP, gamma-amino butyric acid (GABA) permease; madM, malonate transporter; citH, citrate/proton symport; dctA1, dicarboxylate transporter; kgtP, ketoglutaric acid transporter; egl, endoglucanase; cbhA, cellobiohydrolase; f6p, fructose-6-phosphate. Black arrow indicates influx or efflux of nutrients. Red arrow indicates extracellular enzyme secretion. Violet arrow indicates extracellular enzymatic reaction. Blue arrow indicates entry of nutrient into metabolic pathway. Green arrow indicates delivery through TTSS. Cylinder indicates transporter. Plus sign indicates induction of genes.