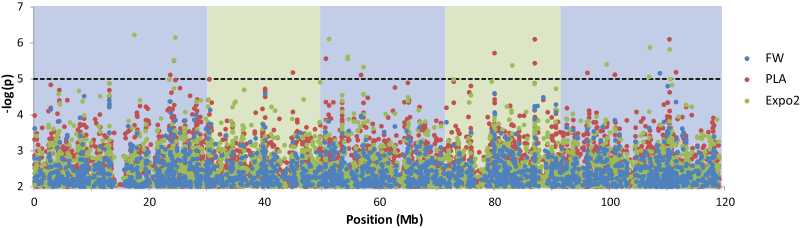
Fig. 5.
Genome-wide association (GWA) mapping of FW, PLA, and parameters of growth model Expo2. Univariate GWA analyses were performed for all traits; in addition, the model parameters ‘r’ and ‘A 0’ were also analysed together in an MTMM-GWA approach. A Manhattan plot of the –log(P) marker–trait association for FW, PLA, and model parameters of Expo2 is shown. PLAs on the different days are represented by one value; for each SNP, only the –log(P) value of the day with the highest association is plotted. Univariate analyses of ‘r’ and ‘A 0’, and the MTMM analyses of ‘A 0’ and ‘r’ jointly are also represented by one value; for each SNP, only the –log(P) value of the analysis with the highest association is plotted. The total number of tested SNP markers was 214 000, but only the ~10 000 SNPs with –log10(P)>2 are plotted. The dotted line indicates the arbitrary threshold of –log(P)=5.