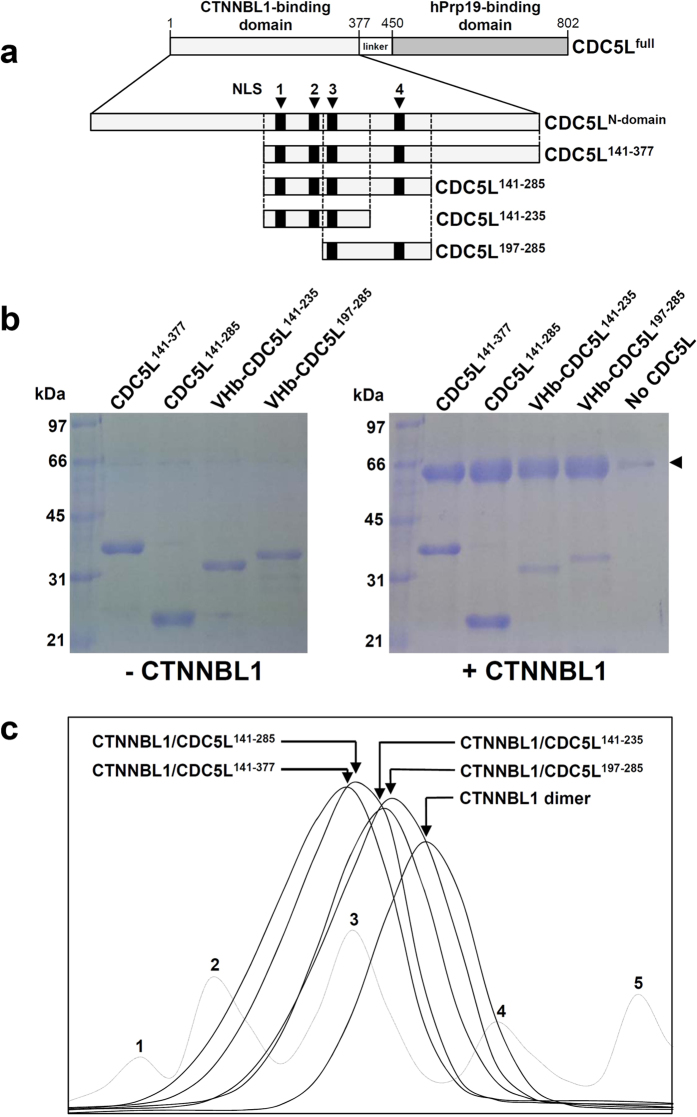
Figure 3. Interaction between CTNNBL1 and four truncated forms of CDC5L.
(a) Construction of four truncated forms of CDC5L. The CTNNBL1- and hPrp19-binding domains of CDC5L are shown, and the residue numbers are indicated. The four NLS-like regions are shown as black boxes and labeled. The CDC5L141–377, CDC5L141–285, CDC5L141–235, and CDC5L197–285 constructs are shown as rectangular boxes. (b) Pull-down assays with CTNNBL1 and the four truncated forms of CDC5L. The left and right images show the SDS-PAGE patterns of the four truncated forms of CDC5L without and with CTNNBL1, respectively. The CDC5L proteins used are indicated at the top of each image. The molecular weights of the standard size markers are indicated on the left side of each image. CTNNBL1 is indicated with an arrow in the image on the right. (c) Size-exclusion chromatography analysis of the CTNNBL1/CDC5L complexes. The elution peaks of the CTNNBL1/CDC5L141–377, CTNNBL1/CDC5L141–285, CTNNBL1/CDC5L141–235, and CTNNBL1/CDC5L197–285 complexes and CTNNBL1 alone are indicated by arrows and labeled. For a precise analysis of molecular weight, standard samples of thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (44 kDa) were used for calibration in size-exclusion chromatography experiments; the peaks are labeled from 1 to 5, respectively.