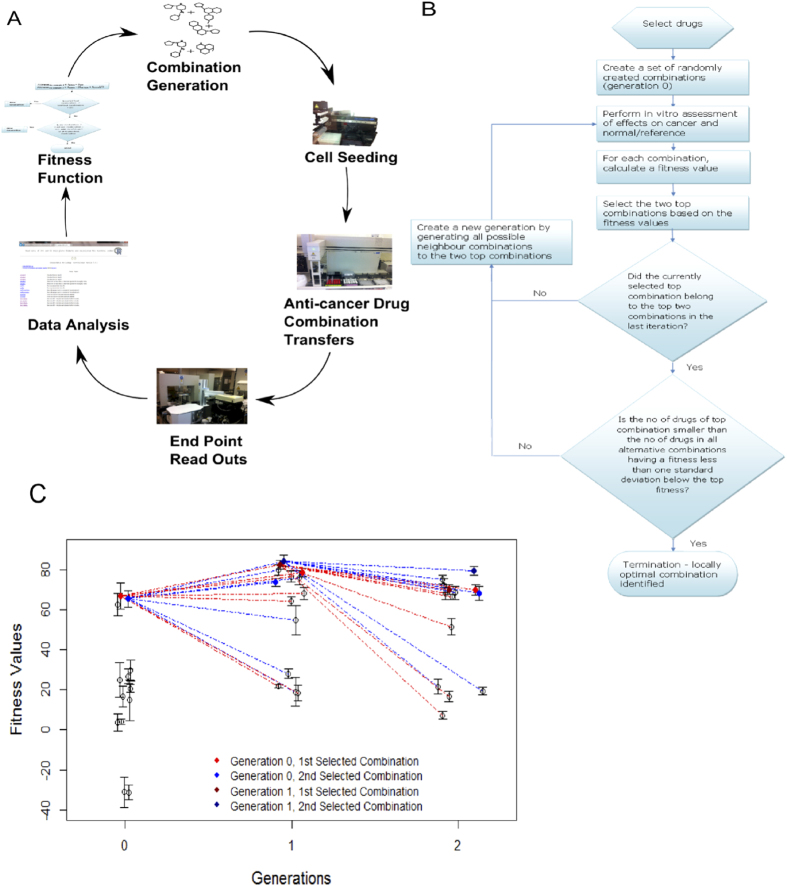
Figure 1.
(A) Iterative search for drug combinations of arbitrary sizes. The procedure starts by generating an initial generation (population) of drug combinations randomly or guided by biological prior knowledge and assumptions. In each iteration the aim is to propose a new generation of drug combinations based on the results obtained so far. The procedure iterates through a number of generations until a stop criterion for a predefined fitness function is satisfied. (B) Overview of the TACS (Therapeutic Algorithmic Combinatorial Screen) algorithm designed and used in this work for iterative search towards promising drug combinations that offer a large TI value. The procedure iterates through a number of generations until a stop criterion is satisfied, taking experimental variability into account. (C) An example run of the TACS algorithm discussed in the main text. Generation 0 is initialised by a random selection of combinations. The drug combinations are assayed for cytotoxicity in three cell line models to determine a proxy for therapeutic benefit. The top two scoring combinations are selected to form the basis of the next generation. Red/blue lines indicate whether the combination was based on the first/second hit in the previous generation. The algorithm finds a set of improved combinations and terminates in two iterations. Notably, the top hit is actually derived from an ancestor that scored second best. One should also note that the two winners from generation 0 (blue and red) are present in all the three generations. Error bars indicate 95% confidence intervals.