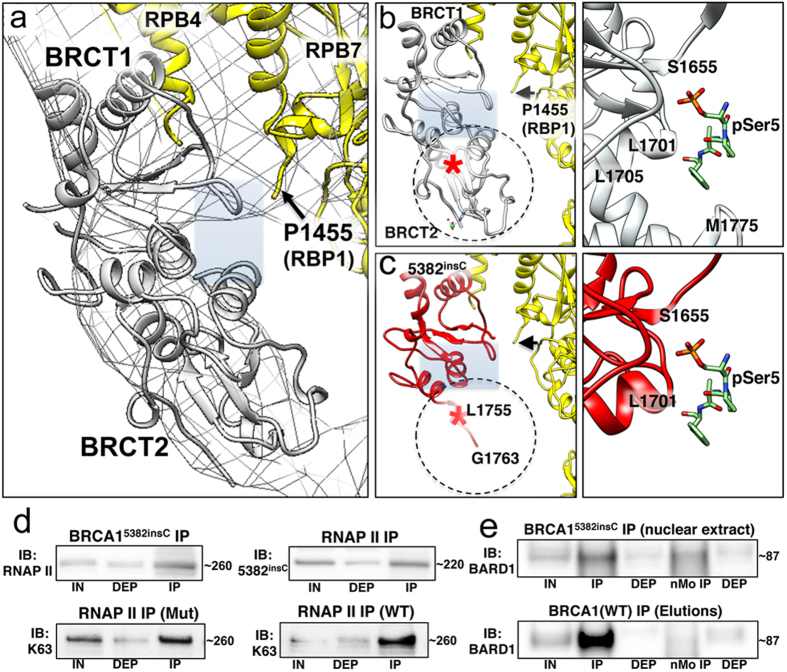
Figure 4. The BRCA15382insC mutation likely alters interactions with RNAP II and BARD1.
(a) Close-up view of the BRCT density with respect to the RPB1 subunit of RNAP II that is disordered beyond P1455 (black arrow), where the C-terminus emanates. Theoretical molecular models of the BRCT domain for wild type BRCA1 (gray; pdbcode, 1KOH) (b) compared to the mutated BRCA15382insC (red) (c) revealed the hydrophobic binding pocket (gray rectangle) is disrupted in the mutated BRCT domain. This significant disruption in the peptide-binding site suggests that native substrates may not interact with BRCA15382insC in the same manner as with wild type BRCA1. (d) Western blot analysis indicates that RNAP II (RPB1 subunit) interacts with BRCA15382insC in co-IP experiments, and the RNAP II core is similarly ubiquitinated by K63-specific moieties in cell lines expressing both mutated (Mut) and wild type (WT) BRCA1. The large subunit of the RNAP II core (RPB1) migrates at ~260 kDa. BRCA1 migrates at ~220 kDa. (e) The BRCA15382insC protein contained in nuclear extracts showed some interaction with BARD1 in comparison to negative control IPs performed using species-specific mouse normal (nMo) IgG antibodies (top panel). Wild type (WT) BRCA1 shows a strong interaction with BARD1 in Ni-NTA eluted fractions used as input material for microchip-capture experiments (bottom panel). BARD1 migrates at ~87 kDa. IN (input material); DEP (unbound); IP (immunoprecipated protein); IB (immunoblot).