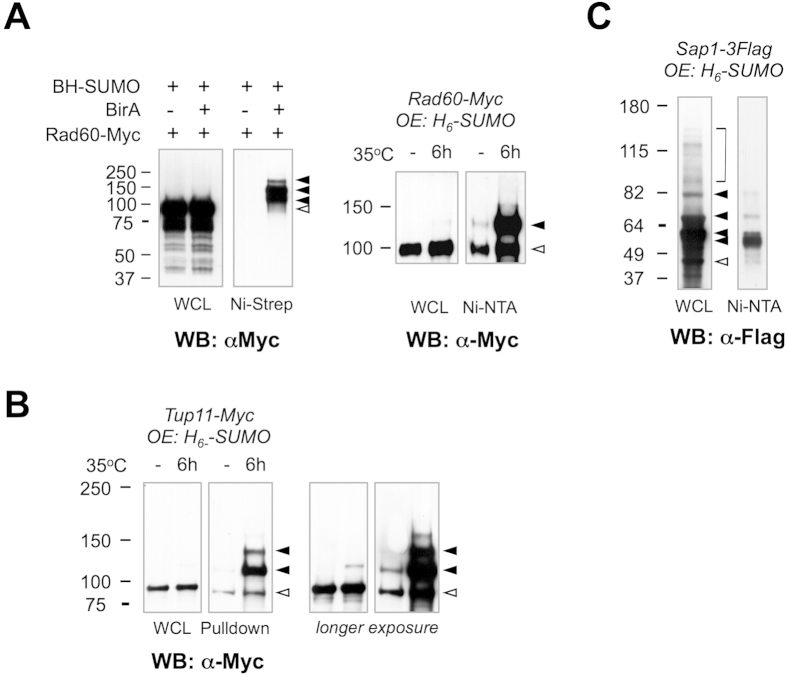
Figure 2. Validation of SUMO targets identified by mass spectrometry.
(A) Left panel: Rad60-Myc and BH-SUMO were expressed from endogenous loci in strains with (+) or without (−) BirA expression. The whole cell lysate (WCL) and eluate after TDAP (Ni-Strep) were analyzed by Western blotting with a Myc antibody. Right panel: A single-step Ni-NTA pulldown was performed using a Rad60-Myc strain that overexpressed (OE) 6-his-tagged SUMO (H6-SUMO) grown at 25 °C, or shifted to 35 °C for 6 h. The WCL and eluate from Ni pulldown (Ni-NTA) were analyzed by anti-Myc Western blotting. (B) Ni-NTA pulldown was performed using an endogenously tagged Tup11-Myc strain overexpressing (OE) H6-SUMO grown at 25 °C, or shifted to 35 °C for 6 h. The WCL and eluate after Ni pulldown (Ni-NTA) were analyzed by anti-Myc Western blotting. A longer exposure is shown on the right. (C) Ni-NTA pulldown was performed on an endogenously tagged Sap1-3xFlag strain overexpressing (OE) H6-SUMO grown at 30 °C. The WCL and eluate after Ni-NTA pulldown were analyzed by anti-Flag Western blotting. In all panels, open triangles denote the position of unmodified protein species, while solid triangles mark the positions of SUMO-modified species. For the pulldown assays, the protein lysate was quantitated by OD280 and equivalent amount of total proteins was used in conditions indicated in the figure.